The mechanisms of DIRAS family members in role of tumor suppressor
Abstract
DIRAS family is a group of GTPases belonging to the RAS superfamily and shares homology with the pro-oncogenic Ras GTPases. Currently, accumulating evidence show that DIRAS family members could be identified as putative tumor suppressors in various cancers. The either lost or reduced expression of DIRAS proteins play an important role in cancer development, including cell growth, migration, apoptosis, autophagic cell death, and tumor dormancy. This review focuses on the latest research regarding the roles and mechanisms of the DIRAS family members in regulating Ras function, cancer development, assessing potential challenges, and providing insights into the possibility of targeting them for therapeutic use.
1 INTRODUCTION
It is well known that cancer remains the leading cause of death, which is due to aggressive characteristics accumulated in biology, diagnosis at a late stage, and inefficient treatment targeted. The cancer progression is a multistep process where cells accumulate a variety of characteristics including sustaining cell proliferation, resisting cell death, evading immunological surveillance, inducing angiogenesis, activating invasion or metastasis, and changing in energy metabolism (Hanahan & Weinberg, 2011).
DIRAS family genes (DIRASs), encoding RAS-related small G-proteins, belonging to a distinct branch of the RAS superfamily (Kontani et al., 2002). In humans, it has been estimated that DIRAS family comprises three functional genes: DIRAS1, DIRAS2, and DIRAS3, dispersed to different chromosomes and tissues (Supporting Information Table S1; Ellis et al., 2002; Kontani et al., 2002; Yu et al., 1999). They have been identified and appear to be associated with a variety of disorders including numerous cancer types. In despite, the high homology to the RAS superfamily members, DIRASs play an extremely different role in regulation of cellular function and development of cancer. DIRASs always compete for invalid binding with RAS to deactivate the downstream effect targets, therefore the RAS signaling pathway (Kontani et al., 2002; Yu et al., 1999). Moreover, DIRASs exert modulatory function by interfering other key signaling pathway associated with cell biology, such as JAK/STAT, FAK, PI-3K/AKT, mTOR, and NF-κB signal transduction (Badgwell et al., 2012; Y. Q. Hu, Si, Ye, Lin, & Zhou, 2013; Lu et al., 2008; Nishimoto et al., 2005; Pei, Yang, Liu, & Qiao, 2011). It is also demonstrated that DIRASs regulate curial effector molecules like calpains and so on, which is essential for regulating cell function and survival (Bao et al., 2002).
In this review, we will concentrate on the role of DIRASs in varied cancers, describe the alterations in cancer hallmarks and signaling pathways that regulate them, and discuss potential therapeutic applications of DIRASs in “DIRAS lost cancers.”
2 DIRAS FAMILY MEMBERS AND THEIR REGULATION OF RAS SIGNALING PATHWAY
The DIRAS family refers to a distinct subgroup of small GTPases, belonging to the RAS superfamily. In structure, DIRAS family numbers share 30–40% amino acid homology with other members of RAS family, featuring five highly conserved domains (G1–G5) mediating guanosine triphosphate (GTP) binding and hydrolysis (Lowy & Willumsen, 1993), a putative effector domain interacting with the downstream effector RAF proteins, and a membrane localizing CAAX motif (where C is cysteine, A is an aliphatic amino acid, and X is any amino acid) at the carboxyl terminus (Kontani et al., 2002; Yu et al., 1999).
Although clearly related in sequence, DIRAS GTPases consist following characteristic substitutions at highly conserved regions among the RAS family: (a) Within the GTP hydrolysis positions, Thr-63 and Ser-65 in DIRAS1 and DIRAS2 are substituted for Ala-59 and Gln-61 in H-RAS, respectively. As for DIRAS3, Ala-46 and Gly-95 replace Gly-12 and Gln-61 in H-RAS, respectively. (b) Within the effector domains, DIRAS proteins have substitutions of Ile-37 (both DIRAS1 and DIRAS2) and Leu-67 (DIRAS3) for Asp-33 in H-RAS. Moreover, Val-40 in DIRAS2 and Asn-72 in DIRAS3 are at a position corresponding to Ile-36 and Asp-38 in H-RAS. (c) Most interestingly, DIRAS3 contains a unique extension of 34 amino acids at the N-terminus, which is required for its growth inhibitory function (Kontani et al., 2002; Luo et al., 2003). Figure 1 shows comparison of amino acid sequences of DIRAS family members and H-RAS.

Comparison of amino acid sequences of human DIRAS family and H-RAS. The amino acid sequences were aligned using Clustal Omega [Color figure can be viewed at wileyonlinelibrary.com]
As predicted by these substitutions, DIRAS GTPases have only a quite low level of GTPase activity and exist predominantly as a GTP-bound form upon its expression in living cells. Moreover, DIRAS GTPases fail to interact with the RAS-binding domain of RAF, resulting in no stimulation of mitogen-activated protein kinase (MAPK; Kontani et al., 2002; Luo et al., 2003; Yu et al., 1999). Furthermore, DIRAS3 was shown to block the RAS–RAF interaction by forming multimeric complex consisting of DIRAS3, C-RAF, and H-RAS, contributing to increased recruitment and anchorage of C-RAF to components of the membrane skeleton, decreased of C-RAF/B-RAF heterodimerization, and C-RAF kinase activity (Baljuls et al., 2012). Additionally, DIRAS3 alternately inhibited RAF activation through an either enhancement homodimerization of KSR1 or recruitment of KSR1 to the RAS-C-RAF complex, and thereby reduce the availability of C-RAF for binding to B-RAF (Baljuls, Dobrzynski, Rauch, Rauch, & Kolch, 2016). DIRAS1 was also proved to antagonize RAS-mediated ERK/MAPK signaling pathway, as diminished ability of RAS to activate Elk-1 in the expression of DIRAS1 (Ellis et al., 2002).
The smg GDP dissociation stimulator (smgGDS) protein is an atypical guanine nucleotide exchange factor that activates many small GTPases, promoting cancer malignancy (Riess, Epplen, Siedlaczck, & Epplen, 1993; Tew et al., 2008). Hence, DIRAS1 can inhibit activity of oncogenic small GTPases and NF-κB transcription in part by nonproductively binding to smgGDS (Bergom et al., 2016). This helps to explain the novel ability of DIRAS family members in antagonization of signaling associated with smgGDS and RAS.
Consequently, distinct from most of oncogenic RAS family members, DIRAS family may tend to be potential tumor suppressor genes (TSGs) in all kinds of cancer types, involved in novel characteristic cellular functions, diverse biological mechanisms, and alterative signaling regulation.
3 DIRASS ACT AS TSGS IN CANCER DEVELOPMENT
Accumulated data have confirmed the negative association between DIRAS family members’ expression and cancer. DIRASs were frequently recognized as a group of TSGs. For instance, DIRAS1 has been considered as a potential human neural tumor suppressor (Ellis et al., 2002). DIRAS2 has been proved to be tightly related to attention deficit/hyperactivity disorder (ADHD), a neurodevelopmental disease with remarkable persistence into adulthood (Reif et al., 2011). DIRAS3 has been identified as the first imprinted putative TSG in adult tumors, with 60% homology to RAS and RAP, expressing consistently in normal ovarian and breast epithelial cells but not in ovarian and breast cancers (Yu et al., 1999). Moreover, induction of DIRAS3 expression significantly decreased fetal bovine serum- and fibronectin-stimulated migration of SKOv3-DIRAS3 ovarian cancer cells when compared with uninduced cells (Badgwell et al., 2012). Re-expression of DIRAS1 and DIRAS2 suppressed growth of human and murine ovarian cancer cells by inducing autophagy-mediated cell death (Sutton et al., 2018).
So dysfunction of DIRASs has been continually discovered in various cancers and exerts its tumor suppressor effects, unlike most of RAS family genes involved in the progression of cancers as pro-oncogenes. Alternative expression of DIRAS family members reported to date as pertaining to different cancer are illustrated in Table 1.
| DIRAS family | Related cancer | Upregulation or downregulation | Regulation patterns | Significant correlated clinicopathological factors | References |
|---|---|---|---|---|---|
| DIRAS1 | Neural tumors | Lost or down | Ellis et al. (2002) | ||
| ESCC | Lost or down | LOH; aberrant methylation |
Advanced clinical stage; lymph node metastasis; shorter survival time |
Zhu et al. (2013) | |
| Colorectal cancer | Lost or down | Aberrant methylation | TNM stage; shorter survival time |
Zheng et al. (2017) | |
| Ovarian cancer | Lost or down | Shorter survival time | Sutton et al. (2018) | ||
| Breast cancer | Lost or down | Bergom et al. (2016) | |||
| DIRAS2 | Ovarian cancer | Lost or down | Shorter survival time | Sutton et al. (2018) | |
| DIRAS3 | Breast cancer | Lost or down | LOH; gene mutation; aberrant methylation and acetylation; miRNA and lncRNA regulation |
Histological differentiation; invasive potential |
Hisatomi, Nagao, Wakita, and Kohno (2002), Y. Li et al. (2013), Peng et al. (2000), Stojic et al. (2016), Wang et al. (2003), Yang et al. (2009), Yu et al. (2003), Yu et al. (1999) |
| Ovarian cancer | Lost or down | LOH; aberrant methylation and acetylation; posttranscriptional regulation |
Shorter survival time | Lu et al. (2006b), Peng et al. (2000), Rosen et al. (2004), Yu et al. (2003), Yu et al. (1999) | |
| HCC | Lost or down | Aberrant methylation | Huang et al. (2009) | ||
| Pancreatic cancer | Lost or down | LOH; aberrant methylation |
Chen et al. (2011), Yang et al. (2010) | ||
| Pancreatic endocrine tumors | Lost or down | Shorter survival time; aggressive progress |
Dalai et al. (2007) | ||
| Gastric cancer | Lost or down | TNM stage; histological differentiation; shorter survival time |
Tang et al. (2012), Wang, Bu, Wang, Zhang, and Zhao (2012) | ||
| Prostate cancer | Lost or down | Aberrant methylation; miRNA regulation | Aggressive progress | Chen et al. (2011), Lin, Cui, Bu, and Yan (2011) | |
| FTC | Lost or down | LOH; aberrant methylation |
Weber et al. (2005) | ||
| Lung cancer | Lost or down | Wu, Liang, Dong, Yu, and Fu (2013) | |||
| Glioma | Lost or down | LOH; aberrant methylation |
Chen, Shi, Yang, and Chen (2014), Riemenschneider, Reifenberger, and Reifenberger (2008) | ||
| Osteosarcoma | Lost or down | Ye et al. (2015) |
- Note. ESCC: esophageal squamous cell carcinoma; FTC: follicular thyroid carcinoma; LOH: loss of heterozygosity; HCC: hepatocellular carcinoma; TNM: TNM classification of malignant tumors.
3.1 DIRAS3 and cancer
Among three GTPases in the DIRAS family, DIRAS3, also called ARIH, is the best investigated in cancer. Yu et al. (1999) reported that DIRAS3 was downregulated in ovarian and breast cancer cells. Meanwhile, low expression of DIRAS3 was proved in clinical cancer tissues compared with corresponding normal tissues. In ovarian cancer, high DIRAS3 expression was found in normal surface epithelial cells, cysts, and follicles, whist low DIRAS3 expression was observed both in low malignant potential tumors and invasive cancers (Rosen et al., 2004). In breast cancer Hisatomi, Nagao, Wakita, and Kohno (2002) found decreased or absent DIRAS3 expression in cancerous tissue samples and the proportion of samples with decreased or absent DIRAS3 expression was associated with the histological stage, indicating that DIRAS3 could be considered as a potential indicator for histological differentiation. DIRAS3 was also downregulated or lost in pancreatic cancers compared with its wide expression in ductal and acinar cells of normal pancreatic tissue (Y. Q. Hu et al., 2013; Lu et al., 2009; Yang et al., 2010). Similar result was found in pancreatic endocrine tumors, that DIRAS3 expression showed a statistically significant difference between either normal pancreas or well-differentiated endocrine tumors and poorly differentiated endocrine carcinomas (Dalai et al., 2007).
Additionally, the frequency of DIRAS3 silencing may be associated with tumorigenesis, aggressive physiological behaviors, and poor prognosis of various kinds of caner types. For example, W. Wang, Bu, Wang, Zhang, and Zhao, (2012) and Tang et al. (2012) found that DIRAS3 protein expression was significantly correlated with tumor differentiation and TNM stage in gastric cancer. It also demonstrated that DIRAS3 expression was further downregulated in invasive carcinoma compared with ductal carcinoma in situ in breast cancer (L. Wang et al., 2003). Lower DIRAS3 expression was also shown to be significantly correlated with cancer aggressive progress (Dalai et al., 2007; Lin, Cui, Bu, & Yan, 2011). All of these studies indicated that DIRAS3 may serve as a good indicator of invasive, metastatic, and aggressive potential in cancer. In addition, patients with lower DIRAS3 expression have been tightly associated with shorter survival time in ovarian, gastric, and pancreatic endocrine cancers (Dalai et al., 2007; Rosen et al., 2004; W. Wang et al., 2012) indicating that DIRAS3 is an valid biomarker for prognosis of cancer.
Furthermore, in contrast to the RAS oncogene, re-expression of DIRAS3 in cancer cells gains the abilities against to cancer hallmarks. It is demonstrated that overexpression of DIRAS3 gene may be associated with inhibition of cell growth (Ellis et al., 2002; Yu et al., 1999) and migration (Badgwell et al., 2012), promotion of apoptosis (Bao et al., 2002), and induction of autophagic cell death and tumor dormancy in carcinomas (Lu et al., 2008). Moreover, DIRAS3 also alters fundamental metabolism in ovarian cancer models which play an increasingly essential role in cancer development. For instance, DIRAS3 expression results in decreased cellular ATP/ADP, increased oxidative stress, and decreased mitochondrial function consistent with programmed necrosis. However, accompanying upregulation of glycolysis and glutaminolysis is autophagy dependent and serves to support cell viability rather than facilitate necroptotic cell death, which may underlie the mechanism of autophagy-dependent dormancy (Ornelas et al., 2016).
3.2 DIRAS1 and cancer
Significant sequence homology between DIRAS1 and DIRAS3 indicates that they may play a similar role in cancer, though there are limited studies available in DIRAS1 and cancer. Recent studies have shown that DIRAS1 protein expression is frequently lost or downregulated in neural tumor-derived cell lines and primary human neural tumors and reduced DIRAS1 protein expression is corresponding to the progress of glioblastoma (Ellis et al., 2002). In human colorectal cancer and primary esophageal squamous cell carcinoma (ESCC), DIRAS1 can induce apoptosis and inhibit cell proliferation, migration, and invasion. Moreover, downregulation of DIRAS1 occurred in those cancers, is significantly associated with advanced clinical stage, lymph node metastasis, and poor overall survival (Y. H. Zhu et al., 2013).
3.3 DIRAS2 and cancer
Previous studies have reported that DIRAS2 plays a fundamental role in the etiology of ADHD and comorbid impulsive disorders (Grünewald et al., 2016; Reif et al., 2011). However, little is known about its functions in cancer. Only recently, together with DIRAS1, DIRAS2 is found to have prognostic value in ovarian cancer. It is also suggested that DIRAS1 and DIRAS2 likely serve as surrogates in the murine genome for DIRAS3, and may function as a backup system to fine-tune autophagy in human (Sutton et al., 2018).
In conclusion, as a putative TSG family, downregulation of the DIRAS family frequently discovered in cancer tissues, plays a pivotal function in cancer pathogenesis and progress and predicts poor prognosis in various tumors. Roles of DIRAS family genes in various cancers and their correlated mechanisms are displayed in Table 2.
| DIRAS family | Biological function | Related cancer | Mechanism | References |
|---|---|---|---|---|
| DIRAS1 | Cell migration inhibition | ESCC; colorectal cancer |
ERK1/2 ↓p38 MAPK ↓ |
Zheng et al. (2017), Zhu et al. (2013) |
| Cell growth inhibition | Neural tumor cells; colorectal cancer; ESCC |
Arrest in cell cycleSmgGDS-mediated DREAM complex ↓SmgGDS-mediated NF-κB ↓ |
Bergom et al. (2016), Ellis et al. (2002), Gonyo et al. (2017), Zheng et al. (2017) Zhu et al. (2013) | |
| Apoptosis induction | ESCC; colorectal cancer |
ERK1/2 ↓p38 MAPK ↓ |
Zheng et al. (2017), Zhu et al. (2013) | |
| Autophagy induction | Ovarian cancer | AKT/mTOR ↓RAS/MAPK ↓ |
Sutton et al. (2018) | |
| DIRAS2 | Cell growth inhibition | Arrest in cell cycleSmgGDS-mediated DREAM complex ↓ |
Gonyo et al. (2017) | |
| Autophagy induction | Ovarian cancer | AKT/mTOR ↓RAS/MAPK ↓ |
Sutton et al. (2018) | |
| DIRAS3 | Cell migration inhibition | Ovarian cancer; gastric cancer; lung cancer; glioma |
JAK/STAT ↓FAK ↓C-RAF/MEK/ERK ↓ |
Badgwell et al. (2012), Chen et al. (2014), Klingauf et al. (2013), Tang et al. (2012), Wu et al. (2013) |
| Cell growth inhibition | Ovarian cancer;breast cancer;pancreatic cancer;prostate cancer;gastric cancer;lung cancer; HCC;glioma; Osteosarcoma |
Arrest in cell cycleAngiogenesis↓Cyclins and CDK ↓CDKI↑PI-3K/AKT/mTOR ↓NF-κB↓ERK1/2↓STAT3↓ |
Chen et al. (2014), Chen et al. (2011), Hu, Si, Ye, Lin, and Zhou (2013), Huang et al. (2009), Lu, Qian, Yu, Yang, and Li (2009), Tang et al. (2012), Wu et al. (2013), Ye et al. (2015), Yu et al. (1999), Zhao, Li, Zhuo, and Cai (2010), Zhu et al. (2014), Zuo et al. (2014) | |
| Apoptosis induction | Ovarian cancer; breast cancer; gastric cancer; colon cancer cell; pancreatic cancers; lung cancer; glioma; osteosarcoma | Calpains ↑PI-3K/AKT ↓NF-κB ↓ |
Bao et al. (2002), Chen et al. (2014), Chen et al. (2011), Hu et al. (2015), Hu et al. (2013), Li et al. (2014), Y. Li et al. (2012), Pei, Yang, Liu, and Qiao (2011), Tang et al. (2012), Wu et al. (2013), Yang et al. (2010), Ye et al. (2015) | |
| Autophagy induction | Ovarian cancer | AIC↑PI-3K/AKT/mTOR ↓AMPK/TSC1/TSC2 ↑RAS/ERK ↓Rab7 ↑;ATG4 ↑;LC3-I ↑ |
Lu et al. (2014a), Lu et al. (2008), Lu et al. (2014b) | |
| Metabolism alteration | Ovarian cancer | Unknown | Ornelas et al. (2016) |
- Note. AIC: autophagosome initiation complex; ESCC: esophageal squamous cell carcinoma; HCC: hepatocellular carcinoma.
4 EXPRESSION REGULATION OF DIRAS FAMILY MEMBERS
As a putative TSG family, expression and function of DIRAS family members are always abrogated in cancer. Exploration of the causes of DIRAS family dysfunction will promote an exhaustive understanding of the causes of DIRAS family-dysfunction-related cancer. It has been determined that frequently genetics changes, epigenetic alterations, transcriptional, and posttranscriptional regulation are common mechanisms for DIRAS family members’ dysfunction. Mechanisms of DIRAS family members’ dysfunction reported to date as pertaining to different cancer are illustrated in Table 1.
4.1 Genetic alteration of DIRAS family members
Intragenic mutations and loss of chromosomal material (loss of heterozygosity [LOH] or homozygous deletion) are two main pathways to disable tumor suppressors (Jones & Laird, 1999). Resided within LOH hotspots, loss of heterozygosity of DIRAS3 and DIRAS1 genes has been frequently detected in ovarian, breast, and follicular thyroid carcinoma (FTC) and ESCC, respectively (Peng et al., 2000; Weber et al., 2005; Yu et al., 1999; Y. H. Zhu et al., 2013). Along with the homozygous deletions, mutations of human DIRAS3 gene occurring at part of the promoter and the exon2 coding region may alter the expression of DIRAS3 in human breast cancer and early-stage lesions, however, concrete mechanism underlying remains unknown (J. Yang et al., 2009).
4.2 Epigenetics alterations of DIRAS family members
Inappropriate gene silencing is alternatively associated with heritable epigenetic changes that involve DNA-methylation and chromatin-remodeling events (Jones & Baylin, 2002; Jones & Laird, 1999). Silencing of DIRAS family members can also be occurred by epigenetics alterations. For example, loss of DIRAS3 expression partly results from acetylation and methylation of chromatin associated with the DIRAS3 promoter, as well as demethylation and inhibition of histone deacetylation can be reactive of both the silenced paternal and imprinted maternal alleles in breast and ovarian cancer cells (Feng et al., 2008; Yu et al., 2003). Among three CpG islands in the DIRAS3 gene, CpG islands II located in the promoter region plays the most important role in epigenetics modification of DIRAS3, as complete loss of DIRAS3 expression resulted from hypermethylation of CpG island II nor did CpG island I (Yuan et al., 2003). Meanwhile, reactivation of DIRAS3 is strongly related to the methylation status of the CpG island II, with a corresponding increase in histone H3 lysine 9/18 acetylation and decrease in histone H3 lysine 9 methylation (Fujii et al., 2003). This mode of DIRAS3 dysfunction is a frequent occurrence in hepatocellular carcinoma, prostate cancer, and oligodendroglioma (Y. Chen et al., 2011; Huang et al., 2009a; Riemenschneider, Reifenberger, & Reifenberger, 2008). In addition, Li et al. (2013a) reported that acetylated STAT3 bound to the DIRAS3 promoter and recruited DNA methyltransferase1 for promoter methylation, accounting for the hypermethylation of the DIRAS3 promoter. Fu et al. (2015) demonstrated the epigenetic repression of DIRAS3 by enhancer of zeste homology 2 (EZH2) through catalyzing trimethylation on H3K27. Similarly, the expression of DIRAS1 is downregulated by promoter region methylation in colorectal cancer and ESCC (Zheng et al., 2017; Y. H. Zhu et al., 2013).
4.3 Transcriptional and posttranscriptional regulation of DIRAS family members
Both the transcriptional and posttranscriptional regulation can contribute to the dramatic decrease in DIRAS3 expression of cancer to some extent. As for transcriptional regulation, crucial transcription factors for the G1/S transition and DNA replication in mammalian cells, E2F1 and 4, bound to the DIRAS3 promoter, reducing DIRAS3 promoter activity two- to three-fold (Lu et al., 2006a). Moreover, recent developments in the field of microRNA (miRNA) and long noncoding RNA (lncRNA) have heightened their crucial role in transcriptional silencing of genes. Y. Chen et al. (2011) and Li et al. (2013b) demonstrated that decreased level of DIRAS3 could be caused by direct targeting of the three prime untranslated region (3′-UTR) of DIRAS3 by miR221/222 in prostate and breast cancer cells. Moreover, lncRNA GNG12-AS1 downregulate the expression of the already active DIRAS3 allele in cis, with diminished Pol II binding and the redistribution of active histone marks at DIRAS3 transcriptional start site (Stojic et al., 2016). In addition to the effect of transcriptional regulation, significantly reduced half-life of DIRAS3 mRNA has been observed in ovarian cancer cells compared with normal ovarian epithelial cells. This rapid turnover of DIRAS3 mRNA probably results from reduced human antigen R (HuR)-AU-rich elements (ARE) binding, whose function is to stabilize mRNAs (Lu et al., 2006b).
Knudson's hypothesis that two hits are required for the full inactivation of a tumor-suppressor gene has been shown to be fundamentally correct in almost all cases of human cancer (Jones & Laird, 1999). Therefore, expression regulation patterns above-mentioned always work together to deactivate DIRAS family members. For examples, homozygous deletions in conjunction with hypermethylation of the genomically imprinted allele lead to complete silencing of the putative maternally imprinted TSG DIRAS3 in ovarian, breast cancers, and FTC (Weber et al., 2005; Yu et al., 1999).
5 HOW DIRAS FAMILY MEMBERS INHIBIT CELL MIGRATION
Cell migration is an exceedingly complex process characterized by cyclical formation and turnover of protrusions and focal adhesions that function both as sites of traction and signaling nexus (Ridley et al., 2003; Romer, Birukov, & Garcia, 2006). Promotion of cell migration during tumorigenesis can be acquired by the activation of extracellular proteases and the altered binding specificities of cell adhesion molecules (CAMs), cadherins, and integrins (Hanahan & Weinberg, 2000). Therefore, interruption of the association of molecular complex within focal adhesions such as signal transducer and activator of transcription 3 (STAT3) and focal adhesion kinase (FAK) contributes to inhabitation of cell migration. Nuclear import of STAT3 is a necessary process for cancer cell progression and metastasis, required for importins and GTP-binding protein Ran (Moore & Blobel, 1993; Terry, Shows, & Wente, 2007). DIRAS3 can bind to STAT3 and importins, respectively, to interrupt their association, thus leading to the defect in nuclear import and transcriptional activity of STAT3 (Figure 2; Nishimoto et al., 2005). Moreover, Muthu, Panneerselvam, Topno, Jayaraman, and Ramadas (2015) implied detailed structural perspective related to the unavailability of Ran-importin β and STAT3 complex. Additionally, DIRAS3 also blocks nuclear localization of other cargo proteins through disrupting the interaction of nuclear localization signal with Ran-importin β and α complexes and competing for importin-Ran binding at the same site with Ran (Huang, et al., 2009b; Nishimoto et al., 2005).
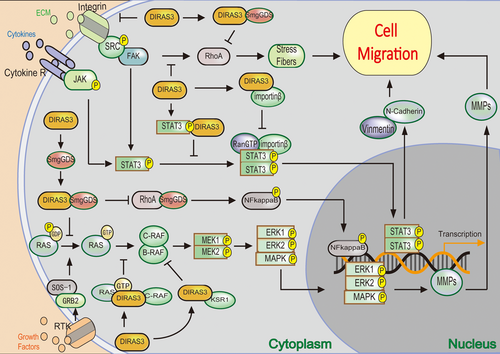
The regulation of DIRAS family members in cell migration and antagonization of smgGDS-associated signaling pathway. (a) DIRAS3 interferes with JAK/STAT signal transduction to suppress migration, through bonding to STAT and preventing its translocation to nucleus. (b) DIRAS3 inhibits the FAK signaling pathway and following RhoA activity and actin stress fiber formation. (c) DIRAS3 prevents smgGDS stimulated RhoA activity and following signaling pathway, through bonding to smgGDS. (d) DIRAS3 interrupts RAS/RAF/MEK/ERK signaling pathway to inhibit migration. FAK: focal adhesion kinase; smgGDS: smg GDP dissociation stimulator; JAK: Janus kinase; STAT; signal transducers and activators of transcription [Color figure can be viewed at wileyonlinelibrary.com]
In summary, re-expression of DIRAS3 inhibits cancer cell migration in following three distinct signaling pathways: (a) Interfering with JAK/STAT signal transduction, through bonding to STAT3 and preventing its localization both in the focal adhesions and nucleus; (b) inhibiting the FAK signaling pathway, resulting in disrupted focal adhesions, decreased RhoA activity and subsequently inhibition of actin stress fiber formation; (c) suppressing C-RAF-MEK-ERK signaling pathway by associating with C-RAF, in contrast to RAS (Figure 2; Badgwell et al., 2012; Klingauf et al., 2013). Meanwhile, It is also reported that DIRAS3-expressing cells inhibit migration and invasion through negatively regulation of integrin β1 and matrix metalloproteinases (MMP)-1/2, which have a well-established role in cell adhesion, motility, and invasion in various types of cancers, including ovarian cancer (Lu & Bast, 2013; Wu, Liang, Dong, Yu, & Fu, 2013).
Similarly, DIRAS1 is proved to act through extracellular signal-regulated kinase (ERK1/2; MAPK3/1) and p38 mitogen-activated protein kinase (MAPK; MAPK14) signaling to trigger MMP-2/9 transcriptional inactivation to inhibit metastasis (Y. H. Zhu et al., 2013).
6 HOW DIRAS FAMILY MEMBERS REGULATE CELL GROWTH
It was first reported that DIRAS3 suppresses growth in ovarian and breast cancer cells (Yu et al., 1999), then the cell growth inhibition capacity of DIRAS3 is discovered in various cancers including hepatocellular carcinoma (HCC; Huang, et al., 2009a), pancreatic cancers (Lu et al., 2009), prostate cancer (Y. Chen et al., 2011), gastric cancer (Tang et al., 2012), lung cancer (Wu et al., 2013), glioma (J. Chen, Shi, Yang, & Chen, 2014), and osteosarcoma (Ye et al., 2015). Mechanically, DIRAS family members exert its growth-inhibitory effects through arrest in cell cycle, decrease in angiogenesis, and changes in signaling pathways. DIRAS3 re-expression has significantly retarded cell growth in vitro and in vivo through blocking cell cycle progression at the G1 phase in breast, ovarian, pancreatic, prostate, lung cancers, HCC, and glioma (Lu et al., 2009; Y. Chen et al., 2011, J. Chen et al., 2014; Wu et al., 2013; Zhao, Li, Zhuo, & Cai, 2010; Q. Zhu et al., 2014; Zuo et al., 2014). Corresponding to cell cycle arrest, modulation of several key G1 regulatory proteins, such as cyclins proteins, cyclin-dependent kinases, and cyclin-dependent kinase proteins inhibitors is found in various cancers. As cyclin D1 is a critical target of proliferative signal in G1, required for G1-S progression (Baldin, Lukas, Marcote, Pagano, & Draetta, 1993), Yu et al. (1999) first demonstrated that DIRAS3 functions as a negative regulator of cell growth in cell culture and heterograft, presumably through downregulation of the cyclin D1 promoter activity and induction of cyclin-dependent kinase proteins inhibitorp21(WAF1/CIP1). Moreover, Lu et al. (2009) proved increase of p21(WAF1/CIP1) and p27kip1, decrease of cyclins A and D1, along with activity reductions of cyclin-dependent kinases 2 (CDK2) and CDK4 in DIRAS3 transfected cells. However, Rosen et al. (2004) revealed that DIRAS3 expression is only correlated with the expression of p21(WAF1/CIP1) but not with cyclin D1 in clinical epithelial ovarian cancers. Furthermore, cyclin-dependent kinase inhibitor 1A (CDNK1A) and growth arrest and DNA-damage-inducible alpha (GADD45A) are upregulated more than five-fold by over expression of DIRAS3 in prostate cancer, which function as regulators in the G1 phase of cell cycle progression (Chen et al., 2011).
Regarding the signaling pathways implicated in the regulation of cell growth, DIRAS3 has been proved to inhibit cell proliferation through blocking PI-3K/AKT, mTOR, NF-κB, and ERK1/2 and STAT3 protein kinases related proliferation signaling pathways (Figure 3; Y. Q. Hu et al., 2013; Lu et al., 2009; Ye et al., 2015; Zhao et al., 2010; Q. Zhu et al., 2014). In addition, it is suggested that mutation or deletion of DIRAS3 protein sequence plays an important role in DIRAS3's inhibitory function. Mutation of the conserved CAAX box at the C-terminus and deletion of the unique N-terminal extension in DIRAS3 nearly abolishes its inhibitory effect on cell growth (Luo et al., 2003).
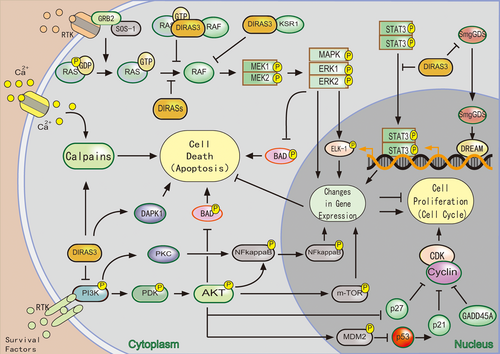
The regulation of DIRAS family members in cell growth and apoptosis cell death. (a) DIRAS3 regulates components of cell cycle control (p21, p27, GADD45A, cyclins proteins, cyclin-dependent kinases) to arrest cell cycle. (b) DIRAS1/2 antagonizes smgGDS-associated proliferative signaling pathway. (c) DIRASs prevent RAS/RAF/MEK/ERK signaling pathway to inhibit cell growth and induce apoptosis. (d) DIRAS3 interferes with PI-3K/AKT, STAT3, and NF-κB signal transduction to inhibit cell growth and induce apoptosis. (e) DIRAS3 induce calpains-dependent apoptosis [Color figure can be viewed at wileyonlinelibrary.com]
Overexpression of DIRAS1 inhibits cell growth and survival in neural tumor, colorectal cancer, and ESCC cells, however the precise molecular mechanisms through which DIRAS1 is involved remain unclear (Ellis et al., 2002; Zheng et al., 2017; Y. H. Zhu et al., 2013). Only recently, DIRAS1 and DIRAS2 have been found to arrest cell cycle by diminishing expression of DREAM target genes and signify nucleolar stress, due to smgGDS depletion in cells (Gonyo et al., 2017). Predicated by similar protein construction, the DIRAS family may function in similar mechanisms to inhibit cell growth, and further research need to be conducted to verify this prospect.
7 HOW DIRAS FAMILY MEMBERS REGULATE APOPTOSIS
Apoptosis (type I programmed cell death) is an important regulatory mechanism that eliminates unwanted cells during development and maintenance of tissue homeostasis, normally through activating poly ADP-ribose polymerase (PARP) and subsequently fragments of effector caspases. However, recent studies have demonstrated existence of attractive candidates calpains for mediating certain events during apoptosis (Johnson, 2000). Calpains are regulated by Ca2+ and target cellular proteins that are instrumental for maintaining the integrity of the cytoskeleton, implicated in apoptosis based on two types of observations: the activation of calpains during cell death and the inhibition of apoptotic execution by various calpain inhibitors (Squier et al., 1999). Instead of normal caspase-dependent apoptosis, re-expression of DIRAS3 in breast and ovarian cancer cells appears to induce apoptosis through a calpain-dependent mechanism followed nucleus accumulation of PARP (Bao et al., 2002; Li et al., 2012). Stimulation of AKT or NF-κB-mediated signaling pathways has been documented to suppress apoptosis in cancer cells (Karin & Lin, 2002; Testa & Bellacosa, 2001). Recently, Pei et al. (2011) and Li et al. (2014) demonstrated that proapoptotic effect of DIRAS3 on cells is involved in the inactivation of PI-3K/AKT survival pathways. Meanwhile, downregulating the NF-κB signaling pathway resulted in the promotion of cell apoptosis (Y. Q. Hu et al., 2013; Pei et al., 2011). Moreover, DAP-kinase 1, caspase-1, cluster of differentiation 70 (CD70), and Bcl-2 interacting protein (HRK) involved in the regulation of apoptosis are reported to be upregulated, corresponding to enhancement of apoptosis (Y. Chen et al., 2011; Wu et al., 2013; H. Yang et al., 2010). However, Washington et al. (2015) reported that re-expression of DIRAS3 fails to induce apoptosis but enhances cisplatin-induced apoptosis, resulting from increasing caspase-3 activation and PARP cleavage by inhibiting ERK and HER2 activity and downregulation of their downstream targets, Bcl-2 and XIAP.
Additionally, ectopic expression of DIRAS1 promotes apoptosis through triggering BAD Ser112 dephosphorylation by stimulating ERK and MAPK signaling pathway (Y. H. Zhu et al., 2013). Regulation of DIRAS family members in apoptosis is displayed in Figure 3.
8 HOW DIRAS FAMILY MEMBERS REGULATE AUTOPHAGIC CELL DEATH AND TUMOR DORMANCY
Autophagy is a “self-digestion” process that can regulate cell survival in a bidirectional way, in response to different contexts and stress levels. Autophagy can recycle damaged cellular organelles and possibly growth factors, as well as maintain chromosome integrity, both contributing to tumor cell death. Meanwhile, autophagy also can help cancer cells to resist anticancer treatments and survive in conditions of low nutrient supply (Amaravadi & Thompson, 2007; Marx, 2006; Mizushima, Levine, Cuervo, & Klionsky, 2008). The cellular events during autophagy follow distinct stages: vesicle nucleation (formation of the isolation membrane/phagophore), vesicle elongation and completion (growth and closure), fusion of the double-membraned autophagosome with the lysosome to form an autolysosome, and lysis of the autophagosome inner membrane and breakdown of its contents inside the autolysosome (Levine & Kroemer, 2008). DIRAS3, required for the induction of both spontaneous and rapamycin-induced autophagy in ovarian cancer cells, can induce autophagic cell death or tumor dormancy in a context-dependent manner by alteration in autophagy hierarchical regulation or execution (Figure 4; Lu et al., 2008).
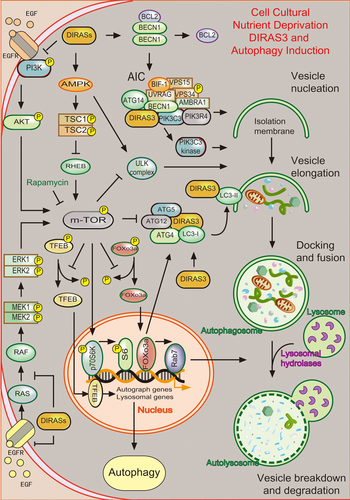
The regulation of DIRAS family members in different steps of autophagy cell death in cell cultural. (a) Nucleation: DIRAS3 → assembly of the AIC (DIRAS3, BECN1, PIK3R4, ATG14, PIK3C) → PIK3C3 kinase activity → vesicle nucleation; DIRASs → PI-3K/AKT↓, AMPK/TSC1/TSC2↑→mTOR↓→ULK complex activity → vesicle nucleation. (b) Elongation: DIRAS3 → ATG4 → LC3 conjugated to Atg5-Atg12 conjugation → LC3-II → vesicle elongation; DIRASs → PI3K/AKT↓, RAS/ERK↓→nuclear retention of FOXo3a → LC3-I↑, ATG4↑→vesicle elongation. (c) Docking and fusion: DIRASs → PI3K/AKT↓, RAS/ERK↓→nuclear retention of FOXo3a → Rab7↑→autophagosome formation → lysosome → autolysosome form. (d) DIRAS1/2 → PI3K/AKT/mTOR↓, RAS/ERK↓→nuclear retention of TFEB → autophagy↑ [Color figure can be viewed at wileyonlinelibrary.com]
During the process of autophagy initial, DIRAS3, an essential composition of the autophagosome initiation complex (AIC), interacts with the BECN1, PIK3C3, and ATG14 core complex to promote the AIC assembly and PIK3C3 kinase activity, required for the initiation of the autophagosome and the induction of autophagy (Lu et al., 2014a). Moreover, DIRAS3 initiates autophagy by inhibition of mTOR activity through blocking PI3K/AKT signaling cascade and upregulating the AMPK/TSC1/TSC2 signaling pathway, combined with the decrease in phosphorylation of p70S6 kinase and ribosomal proteinS6 (Lu et al., 2008).
During the process of vesicle elongation and completion, ATG4 associates with microtubule-associated protein 1 light chain3 (LC3) and cleaves LC3-I to produce LC3-II. Sequentially, LC3-II docks to autophagosome membrane to facilitate autophagosomes maturation (Mariño & López-Otín, 2004). Therefore, DIRAS3 facilitates autophagosome formation probably by upregulating ATG4 and LC3-I or directly colocalizing with the elongation marker ATG12, and the maturation marker punctate LC3 (Lu et al., 2008). Moreover, induction of ATG4 and LC3-I can be acquired by nuclear retention of FOXo3a, resulting from downregulation of PI-3K/AKT and RAS/ERK signaling through enhancing internalization and degradation of the epidermal growth factor receptor by DIRAS3 (Lu et al., 2014b).
During the fusion of autophagosomes with lysosomes, DIRAS3-mediated nuclear retention of FOXo3a also increases the expression of Rab7, required for fusion of autophagosomes with lysosomes (Lu et al., 2014b).
Interestingly, instead of inducing autophagic cell death in culture, over expression of DIRAS3 enables tumor cells to remain dormant in mice xenografts or in the presence of survival factors of microenvironment (Lu et al., 2008). Moreover, DIRAS3-mediated autophagy facilitates survival of dormant ovarian cancer cells that remain after conventional chemotherapy (Lu et al., 2014a). This may account for the recurrence of tumor after conventional surgery and chemotherapy, whose underpinning mechanisms were poorly understood, causing huge challenge to recovery of cancer.
Similarly, it is found that DIRAS1 and DIRAS2 induce and regulate autophagy by inhibition of the AKT/mTOR and RAS/MAPK signaling pathways and modulating nuclear localization of autophagy-related transcription factors FOXO3/FOXO3A and TFEB (Sutton et al., 2018).
9 CLINICAL APPLICATION IN DIAGNOSIS AND THERAPEUTICS
As mentioned above, statistic-altered expression of the DIRAS family between cancer and normal tissues makes it possible to apply DIRAS family numbers as novel biomarkers in the area of clinical cancer detection and diagnosis. Significant correlation between focused expression of DIRAS family numbers and good survival of patients indicates DIRAS family numbers may act as novel predictors of preferable prognosis and response to therapy. In conclusion, roles of DIRAS family numbers played in a host of cancer types have opened a way to innovative diagnostic and therapeutic strategies.
Recent advances in realizations of DNA methylation markers in molecular diagnostic and therapeutic applications attest to the great promise of early detection, diagnosis, and therapy of DNA methylation involved cancer. For example, cancer-specific DNA methylation patterns can be detected in tumor-derived free DNA in the bloodstream and in epithelial tumor cells shed into the lumen, offering a promising approach to the early detection of cancer (Laird, 2003). Thus, detection of the specific DNA methylation patterns in the DIRAS family genes can provide a possible way to detect cancer at an early stage. Interestingly, it is manifested that DIRAS3 promoter methylation is a highly significant predictor of survival in patients who had not received tamoxifen therapy but showed no predictive value for patients treated with tamoxifen therapy, indicating its potential indicative role of clinical hormone therapy in breast cancer (Widschwendter et al., 2004). Meanwhile, aberrant methylation of DIRAS3 is associated with negative estrogen and progesterone receptor status, which are indicators of cancer cell growth rate and determine response to endocrine therapies (Barrow et al., 2015). However, the diagnostic and therapeutic potential of DNA methylation profiles is still largely untapped because of the complexity of varying distributions of DNA methylation profiles between cancer and normal tissues.
According to roles that DIRAS family members played in cancer, therapeutic implements to regain DIRAS family members in cancer cells can provide prospects onto promising treatment of DIRAS family lost cancer and the reactivation of DIRAS family members in these cancer cells may serve as new biomarkers with which to monitor the treatment effects. For example, consistent with epigenetics modification, CpG demethylating agents, and histone deacetylase inhibitors are common treatments to regain DIRAS family members in cancer cells (Feng et al., 2008; Fujii et al., 2003). Moreover, a potential nontoxic chemopreventive agent, Genistein, restores expression of DIRAS3 by regulation of miR-221 and miR-222 for treating prostate cancer (Y. Chen et al., 2011). It is anticipated to regain DIRAS family by directly importing genes into cells of patients, which recognized as gene therapy. However, clinical application of this medication exists huge challenges and still has a long way to go. Collaboratively, existent chemotherapy combined with treatment of agents to regain DIRAS3 can exert an assistant effect. It is proved that re-expression of DIRAS3 enhances the cytotoxic effect of cisplatin and paclitaxel in cell culture (Washington et al., 2015; Zou et al., 2011).
To conclude, detection and diagnosis with DIRAS family members, treatment to induce DIRAS family members in patients’ cells, and measure to prevent their loss deserve further explore and validation.
10 CONCLUSION AND FUTURE VISION
We have sought here to synthesize the advances in the DIRAS family and its roles in cancer, which have contributed to form an elementary framework for understanding the complex biologic changes induced by the DIRAS family and investigating the underlying mechanisms associated with cancer. Consistent with similar structure, the tumor suppressive role and biologic mechanical discoveries of DIRAS3 provide a conceptual way for further research of DIRAS1 and DIRAS2 in the foreseeable future. Furthermore, the complexing roles of DIRAS family members in cancer may offer an alternative way to overcome the challenges in the way of fighting against cancer, in combination with more effective strategies targeted to the cancer characteristics and hence the patients.
Looking ahead, we envision continuing elucidation of cancer pathogenesis, including precise controlling mechanics and signaling pathways, to make most utility of the DIRAS family. Meanwhile, we anticipate the subsequent finding of other TSGs recognized by comparable structure. Moreover, consistent exploration of the specific discrepancy between the DIRAS family genes and normal oncogenic RAS genes can help to explain how oncogenic superfamily members acquire tumor suppressive ability. This investigation is going to contribute a lot to the understanding of the conversion between oncogenes and TSGs, and thus to make it possible to transform oncogene into TSG with artificial intervention.
However, restricted in the development of medical technology, clinical treatment targeting DIRASs in curing cancer remains theoretical, which continues to be a matter of challenge and deserves further significant efforts.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation, China (Grant Nos. 81372622 and 81672362); the Zhejiang Provincial Natural Science Foundation, China (Grant No. LQ17H160009); and the Zhejiang Province Medical Science and Technology Project (Grant No. 2018276694).
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.