TNF-α/TNFR2 Regulatory Axis Stimulates EphB2-Mediated Neuroregeneration Via Activation of NF-κB
Abstract
HIV-1 infected individuals are at high risk of developing HIV-associated neurocognitive disorders (HAND) as HIV infection leads to neuronal injury and synaptic loss in the central nervous system (CNS). The neurotoxic effects of HIV-1 are primarily a result of viral replication leading to the production of inflammatory chemokines and cytokines, including TNF-α. Given an important role of TNF-α in regulating synaptic plasticity, we investigated the effects of TNF-α on the development of neuronal processes after mechanical injury, and we showed that TNF-α treatment stimulates the regrowth of neuronal processes. To investigate transcriptional effects of TNF-α on synaptic plasticity, we analyzed both human neurosphere and isolated neuronal cultures for the regulation of genes central to synaptic alterations during learning and memory. TNF-α treatment upregulated Ephrin receptor B2 (EphB2), which is strongly involved in dendritic arborization and synaptic integrity. TNF-α strongly activates the NF-κB pathway, therefore, we propose that TNF-α-induced neurite regrowth occurs primarily through EphB2 signaling via stimulation of NF-κB. EphB2 promoter activity increased with TNF-α treatment and overexpression of NF-κB. Direct binding of NF-κB to the EphB2 promoter occurred in the ChIP assay, and site-directed mutagenesis identified binding sites involved in TNF-α-induced EphB2 activation. TNF-α induction of EphB2 was determined to occur specifically through TNF-α receptor 2 (TNFR2) activation in human primary fetal neurons. Our observations provide a new avenue for the investigation on the impact of TNF-α in the context of HIV-1 neuronal cell damage as well as providing a potential therapeutic target in TNFR2 activation of EphB2. J. Cell. Physiol. 231: 1237–1248, 2016. © 2015 Wiley Periodicals, Inc.
TNF-α is a proinflammatory cytokine that has many important physiological and pathological roles in a broad range of biological events and induces the production of other proinflammatory cytokines and chemokines, as well as itself (Chu, 2013). TNF-α triggers the activation of two major signaling pathways, Ikappa-B (IκB) kinase (IKK)/NF-κB and mitogen-activated protein kinase (MAPK)/AP-1, that regulate the expression of proinflammatory cytokines and induction of many downstream processes, including apoptosis and necrosis (Baud and Karin, 2001). TNF-α signals through two receptors, TNFR1 and TNFR2, that can either be soluble or membrane bound. TNFR1 is ubiquitously expressed, but TNFR2 is mainly present on endoepithelial cells and lymphocytes (Chu, 2013). In the brain, both receptors are expressed by neurons and glial cells, but their distribution is dependent on apoptotic signaling or inflammatory cascades (Kinouchi et al., 1991; Tchélingérian et al., 1995; Botchkina et al., 1997; Dopp et al., 1997; Sairanen et al., 2001; Figiel and Dzwonek, 2007). It has been reported that the differences in their signaling have opposite effects on neurons, with TNFR1 having a damaging impact on neuronal cells whereas TNFR2 appears to have a neuroprotective effect (Fontaine et al., 2002; Marchetti et al., 2004). However, it has been speculated that the damaging effect may be a benefit to neurons in that TNFR1 signaling for apoptosis may reduce the inflammatory cascade and prevent further damage to the surviving neurons (Probert, 2015). TNF-α levels have been shown to be elevated in the brain in response to multiple diseases, including HIV (Fiala et al., 1997; Saha and Pahan, 2003; Zou and Crews, 2005).
TNF-α signaling through NF-κB has various roles in the brain, including neuroprotection during neuronal development mostly in proliferation and migration of neural progenitor cells, dendritic arborization, axonal growth, and regulating synaptic gene expression (Memet, 2006; Kaltschmidt and Kaltschmidt, 2009). Various neurotransmitters and neurotrophic factors and cytokines allow constitutively active NF-κB to persist. In some CNS neurons, excitatory amino acids produce stimuli that also contribute to the constitutive activation of NF-κB (Pizzi and Spano, 2006). NF-κB also has multiple roles in HIV-1 pathogenesis, as it is well demonstrated by in vitro studies and genome-wide analysis that HIV-1 transcription can be initiated by prototypical NF-κB p50 /p65 heterodimers and components of the canonical NF-κB pathway (Quivy and Van Lint, 2004; Calao et al., 2008).
Eph receptors (Ephs) and ephrin receptor ligands (ephrins) are involved in a variety of developmental processes such as cardiovascular and skeletal development, axon guidance, and tissue patterning and are expressed in nearly all tissues of the developing embryo. They have recently been shown to be involved in learning and memory, in bone homeostasis, and in insulin secretion. Alterations of Eph/ephrin signaling in humans have been implicated in congenital diseases and cancer (Arvanitis and Davy, 2008).
Forming the largest subfamily of receptor tyrosine kinases (RTKs), Ephrin receptors interact with cell surface-bound ligands known as ephrins. There are structural differences that define the ligands into two groups: ephrin A (A1–A6) ligands are tethered to the plasma membrane via a glycosylphosphatidyl inositol moiety, and ephrin B (B1–B3) ligands span the plasma membrane and possess a short cytoplasmic tail. The Eph receptors also form two groups based on their affinity for their ligands: EphA (A1-8 and A10) receptors interact with ephrin A (A1-5) and EphB (B1-4 and B6) receptors interact with ephrin B (B1-3). Eph/ephrin signaling demonstrates a unique ability for both, the receptors and ligands, to transduce a signaling cascade upon interaction. The ligand binding to the receptor is considered forward as the signaling proceeds through the intracellular domain of the receptor, and reverse signaling occurs when the receptor binds to the ligand, leading to signaling cascades occurring through the intracellular domain of the ligand. Eph receptor and ephrin interactions can happen in trans between two opposing cells, which is considered activating, or in cis within the same cell, which is commonly regarded as inhibiting (Arvanitis and Davy, 2008). The Eph/ephrin B family of interactions have recently emerged as major role players in synaptic plasticity and neuronal process development. Studies have demonstrated that EphB2 receptor signaling pathways are required for spine morphogenesis, supporting its role in NMDA-dependent synaptic plasticity in the hippocampus, such as long-term potentiation (LTP) and long-term depression (LTD) (Grunwald et al., 2001; Henkemeyer, 2003; Penzes et al., 2003; Murai and Pasquale, 2004).
The role of EphB2 in neurocognitive disorders has been of recent focus, mainly in Alzheimer's disease (Lacor et al., 2007; Cissé et al., 2011). EphB2 levels are altered in HIV affected individuals (Yuferov et al., 2013) but there has not been a conclusive study to correlate EphB2 to cognitive decline that is observed in the neurodegenerative processes of HIV-associated neurocognitive disorders (HAND). In this study, we sought out to determine the potential effects of TNF-α for neurons, particularly on neurite regeneration. We first subjected neurons to induced injury to observe the impact of TNF-α on the ability of the neurites to regrow. TNF-α treatment promoted longer and more complex neurite regrowth in human primary fetal neurons following induced injury, so we next investigated the effects of TNF-α treatment on expression of genes involved in extension of neuronal processes. Among affected genes, EphB2 was of main interest. We found that the expression of EphB2 is stimulated by TNF-α in an NF-κB-dependent pathway. By site mutagenesis, we determined NF-κB binding sites on the EphB2 promoter most responsible for this activation. Finally, TNFR2 was identified as the main receptor for TNF-α-NF-κB-mediated stimulation of EphB2. Additional studies are warranted for better understanding beneficial effects of TNF-α in the CNS (Probert, 2015).
Materials and Methods
Mixed neuronal population culture
Fetal brain tissue (gestational age 16–18 weeks) was obtained from elective abortion procedures performed in full compliance with National Institutes of Health and Temple University ethical guidelines. The experiments were performed in triplicate with different fetal tissue from different donors for each experiment in single passage. The tissue was washed with cold Hanks balanced salt solution (HBSS) and meninges and blood vessels were removed. For primary neuronal isolation, tissue in HBSS was digested with papain (Sigma–Aldrich, St. Louis, MO) for 30 min at 37°C. The tissue was further dissociated to obtain single-cell suspensions by repeated pipetting. Cells were plated at a density of approximately 1.8 × 106 cells/60 mm dish coated with poly-D lysine in Neurobasal media with B27 supplement, horse serum, and gentamicin (Invitrogen, Waltham, MA). After approximately 2 hrs, neurons were re-fed with the same Neurobasal media. Twenty-four hours later, cultures were re-fed with a complete change of Neurobasal media without horse serum. Cells were maintained in Neurobasal medium containing Glutamax, B27 supplement, and gentamicin with half-media changes every three days. Cell types present in cultures were assessed by immunolabeling for cell-type specific markers: TUJ1 (1:200, Millipore, Billerica, MA), Nestin (1:200, BD Biosciences, Franklin Lakes, NJ), and GFAP (1:200, Millipore, Billerica, MA).
Human primary fetal neuronal culture
Fetal brain tissue (gestational age 16–18 weeks) was obtained from elective abortion procedures performed in full compliance with National Institutes of Health and Temple University ethical guidelines. Human Primary Fetal Neuronal Culture was prepared as described previously (Darbinyan et al., 2013a,2013b). The experiments were performed in triplicate with different fetal tissue from different donors for each experiment in single passage. Briefly, the tissue was washed with cold Hanks balanced salt solution (HBSS) and meninges and blood vessels were removed. For primary neuronal isolation, tissue in HBSS was digested with papain (Sigma–Aldrich) for 30 min at 37°C. The tissue was further dissociated to obtain single-cell suspensions by repeated pipetting. Cells were plated at a density of approximately 1.8 × 106 cells/60 mm dish coated with poly-D lysine in Neurobasal media with B27 supplement, horse serum, and gentamicin (Invitrogen, Waltham, MA). After approximately 2 hrs, neurons were re-fed with the same Neurobasal media. Twenty-four hours later, cultures were re-fed with a complete change of Neurobasal media without horse serum. Four days later, half of the media was removed and replaced with Neurobasal media supplemented with fluoro-deoxyuridine (FDU) and uridine. Following FDU treatment, neurons were maintained in Neurobasal medium containing Glutamax, B27 supplement, and gentamicin with half-media changes every three days. Purity of cell type specific cultures was assessed by immunolabeling for cell-type specific markers: βIII Tubulin (1:200, Millipore, Billerica, MA).
Scratch assay
The scratch assay was performed as described by Liang et al. (2007). Human primary fetal neuronal cultures were grown on 6-well plates or 2-chamber glass slides coated with poly-d-lysine as described above. After 14 days in culture, monolayers were scraped in a straight line to create a “scratch” with a sterile plastic cell lifter. Debris was removed and cultures were washed once with 1 ml of the neuronal media and then replaced immediately with neuronal media with no treatment for control conditions or with recombinant TNF-α (Cell Signaling, Danvers, MA) at a concentration of 100 ng/ml. Cells were treated every 4 h for 4 days after initiation of the scratch and ultimately fixed and immunolabeled for immunocytochemical studies as described below.
Cell harvest
Following treatment, cells were washed in 1× PBS, then scraped from plate and collected in ice-cold PBS. Cell suspension was centrifuged at 13,000g for 1 min and the supernatant was discarded. The cell pellet was then re-suspended in ice-cold RIPA lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitor cocktails (Calbiochem, San Diego, CA) followed by rotation at 4°C for 1 h to complete cell lysis. Samples were then centrifuged at 13,000g for 15 min to separate insoluble material. The supernatant containing protein was collected and placed into clean, pre-chilled Eppendorf tubes and samples were stored at −80°C until protein analysis was performed. For experiments requiring nuclear/cytoplasmic fractionation, cells were collected similarly but lysed and fractionated using the NE-PER nuclear and cytoplasmic extraction kit according to the manufacturer's protocol (Thermo-Scientific, Waltham, MA). Following fractionation, nuclear, and cytoplasmic lysates were stored at −80°C until analysis.
Plasmids
The EphB2 promoter region was cloned to contain 840 base pairs upstream from the transcription start site and 83 base pairs downstream of the transcription start site using the following primers adapted from Fu et al. (2009): Forward: 5′-GGGTACCGGGTCTGCCTGCAAGGGC-3′ Reverse: 5′-CCCAAGCTTTCACCTTCCACGGCGGCGAGCAG-3′. The forward primer contains the KpnI restriction site and the reverse primer contains the HindIII restriction site. Following subcloning into the pCR™2.1 vector with the TA Cloning® Kit (Invitrogen, Waltham, MA), the promoter sequence was removed from the pCR™2.1 vector using KpnI and HindIII digestion and ligated into the pGL3-Basic vector (digested with KpnI and HindIII) (Promega, Madison, WI). Positive clones were confirmed via sequencing.
Synaptic plasticity array
Following treatment, cells were harvested in PBS as described above. Total RNA was extracted using the Qiagen RNeasy® kit according to manufacturer's instructions (Qiagen, Valencia, CA) with on-column DNase I digestion according to manufacturer's instructions (Qiagen, Valencia, CA). Complementary DNA (cDNA) was generated using the RT2 First Strand Kit supplied with the RT2 Profiler PCR synaptic plasticity array following the manufacturer's protocol Qiagen, Valencia, CA). After first strand synthesis, samples were added to the RT2 SYBR Green Mastermix and aliquoted to the 96-well RT2 Profiler PCR Array, and the qRT-PCR was performed on a Roche LightCycler® 480 (Roche, Indianapolis, IN). Results were confirmed using quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) with specific primers for the desired target genes as described below.
RNA extraction and quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR).
Following treatment, cells were harvested in PBS as described above. Total RNA was extracted using the Qiagen RNeasy® kit according to manufacturer's instructions (Qiagen, Valencia, CA) with on-column DNase I digestion according to manufacturer's instructions (Qiagen, Valencia, CA). Complementary DNA (cDNA) was generated using the M-MLV Reverse Transcriptase cDNA synthesis kit following the manufacturer's protocol (Invitrogen, Waltham, MA). qRT-PCR analyses of EphB2, GRIP1, and NF-κB were then performed. Primer sequences utilized were: EphB2 (F: 5′-CGTGTTTGAGTCAAGCCAGA-3′; R: 5′-GCTGCAATGGTATCCACCTT-3′), GRIP1 (F: 5′-GTCCTTAGCCTCCAGCAG-3′; R: 5′-ATGGCCATCACTCTGTCTCC-3′), NF-κB (5′-GCACGACAACATCTCATTGG-3′; R: 5′-TCCCAAGAGTCATCCAGGTC-3′), and β-actin (F: 5′-CTACAATGAGCTGCGTGTGGC-3′; R: 5′-CAGGTCCAGACGCAGGATGGC-3′). Reaction mixtures for qRT-PCR consisted of LightCycler® 480 SYBR Green I Master Mix (Roche, Indianapolis, IN), forward and reverse primers for EphB2, GRIP1, NF-κB, or β-actin(housekeeping), 2µL of cDNA (diluted 1: 10) and H2O to bring the reaction volume up to 20 µl. Samples were placed in a 96-well plate and the qRT-PCR was performed on a Roche LightCycler® 480 (Roche, Indianapolis, IN). All reactions were performed in triplicate and amplification curves and p values were obtained for analysis. Results were normalized against β-actin expression as a control housekeeping gene.
Immunocytochemistry
Following the initiation of the scratch assay and appropriate treatment, cells plated on chamber slides were washed once with 1× PBS and fixed with 4% formaldehyde for 15 min at 23°C. Following fixation, slides were washed with 1× PBS and cells were permeabilized with 0.2% Triton X-100 for 15 min, washed again, and placed in blocking solution (5% normal goat or horse serum; Vector Laboratories, Burlingame, CA) for 30 min. Slides were then incubated in primary antibody for 2 hrs at 23°C. Primary antibodies consisted of Nestin (1:200; BD Biosciences, Franklin Lakes, NJ), TUJ1 Alexa Fluor-labeled (Covance, Berkeley, CA), GFAP (1:200; Santa Cruz Biotechnology, Dallas, TX), βIII Tubulin (1:200, Millipore, Billerica, MA), or EphB2 (1:200; Cell Signaling, Danvers, MA). Following incubation with primary antibody, slides were rinsed 3× with PBS, and incubated with fluorescein isothiocyanate (FITC) (1:500; Vector Laboratories, Burlingame, CA) or Texas Red (1:500; Vector Laboratories, Burlingame, CA)-conjugated secondary antibodies for 1 h at room temperature in the dark. Sections were washed 3× with PBS, cover-slipped with an aqueous based mounting medium containing DAPI for nuclear labeling (Vectashield; Vector Laboratories, Burlingame, CA), and visualized with Leica Advanced Widefield imaging system (Leica Microsystems; Buffalo Grove, IL).
Chromatin immunoprecipitation (ChIP) assay
The chromatin immunoprecipitation (ChIP) assay was carried out following the Millipore (Billerica, MA) protocol. Human primary fetal neurons were cultured as described and allowed to grow for 14 days. Cells were washed twice with PBS and cross-linked with 1% formaldehyde at room temperature for 10 min. After cross-linking with formaldehyde, treated cells were lysed in SDS lysis buffer (1% SDS, 10 mM EDTA and 50 mMTris, pH 8.1) containing 1× protease inhibitor cocktail (Calbiochem, San Diego, CA) and sonicated such that DNA fragments were 200–1,000 base pairs in length. Supernatants were collected and diluted in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl) followed by immunoclearing with protein A-agarose beads saturated with salmon sperm DNA (Millipore, Billerica, MA) for 2 h at 4°C. Extracts were then incubated overnight with 1 µg/ml of NF-κB p65 antibody (Santa Cruz, Dallas, TX) or rabbit IgGs at 4°C with end-over-end rotation. After incubation, 50µL of Protein-A bead slurry (Millipore, Billerica, MA) were added and samples were incubated with end-over-end rotation at 4°C for 1 h The beads were then washed once with each of the following buffers in successive order: Low Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), High Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), and LiCl Immune Complex Wash Buffer (0.25 M LiCl, 1% IGEPAL CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris, pH 8.1). The beads were then washed with 1× TE twice and the histone complex was eluted using elution buffer (1% SDS, 0.1M NaHCO3). Cross-links were reversed at 65°C for 4 hrs using 20 μL 5 M NaCl, protein digestion was performed using 10 μl 0.5 M EDTA, 20 μl 1M Tris-HCl (pH 6.5), and 2 μl 10 mg/ml Proteinase K for 1 h at 45°C. Precipitated DNA was purified using phenol/chloroform extraction. For PCR, 1 µl from a 50 µl DNA extraction and 25 cycles of amplification were used. NF-κB sites of the EphB2 promoter region were amplified using the following primers:
EPHB2 NF-κB site 1: F: 5′-GTCGCCGCGCTCCAG-3′; R: 5′-GCACGGCCCACCTGAG-3′; NF-κB site 2: F: 5′-AGTTCGGTCCCTTTCGAAGC-3′; R: 5′-GAATGCTGCAAAGCCGGAG-3′; NF-κB site 3: F: 5′-AGCTCAGAACAACGGGGC-3′; R: 5′-GCTTCGAAAGGGACCGAACT-3′; IκBα F: 5′-GACGACCCCAATTCAAATCG-3′; R: 5′-TCAGGCTCGGGGAATTTCC-3′.
PCR products were analyzed on 2% agarose gels and sequencing confirmed amplification of predicted NF-κB sites.
Luciferase reporter assay
Human primary fetal neurons were plated at 80% confluency and 14 days later were transfected with the pGL3-EphB2-luciferase reporter plasmid, containing the human EphB2 promoter region (−840 to +83 base pairs in relation to the transcription start site) driving a luciferase reporter gene using Lipofectamine® 2000 (Thermo Fisher Scientific, Waltham, MA). After 24 hrs. the treatment with TNF-α was repeated. The cells were harvested 30 min later and protein extracts were prepared, 20 μg of which were used to examine the level of luciferase activity using the Luciferase assay reagent (Promega, Madison, WI). Relative luciferase units were obtained for each sample and normalized to total protein concentration.
Point mutation analysis
Human primary fetal neurons were cultured in the same manner as described in Luciferase Reporter Assay. Before the transfection was performed, the reporter plasmid was mutated for NF-κB binding sites 2 and 3 to prevent binding using the QuikChange II-XL Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Agilent Technologies, Santa Clara, CA) using the following primers to direct mutagenesis:
EPHB2 NF-κB site 2: F: 5′-CCCCCGCTTTCTCAAGCCCCTTCCCCGCC-3′; R: 5′-GGCGGGGGAAGGGGCTTGAGAAAGCGGGGG-3′; NF-κB site 3: F: 5′-CCCCGGTGCCTCTCAAAGCCGCCTGTTTTG-3′; R: 5′-CAAAACAGGCGGCTTTGAGAGGCACCGGGG-3'.
After confirming successful mutagenesis via sequencing, cells were transfected and luciferase assay was performed as described above.
TNFR blocking experiment
Human primary fetal neurons were plated at 80% confluency and 14 days later, treatment was initiated to block either TNFR1 or TNFR2 using either TNFR1 or TNFR2 neutralizing antibodies (Santa Cruz, Dallas, TX) at a concentration of 2 μg/ml with IgG antibody treatment as the nonspecific control (2 μg/ml). After a 30 min incubation, TNF-α treatment was initiated at a concentration of 100 ng/ml for 24 hrs and again 30 min prior to harvesting RNA. qRT-PCR was performed for EphB2 expression exactly as performed for previous experiments described in qRT-PCR.
Results
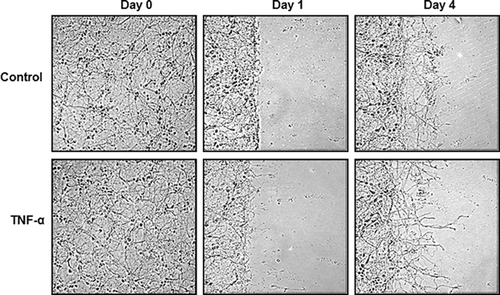
TNF-α treatment induces longer and more complex neurite outgrowth upon mechanical injury in in vitro scratch assay
The effects of TNF-α on neurite regrowth were first investigated considering the dual role of TNF-α. For this, the migration, “scratch,” assay for a model of induced injury was adapted and human primary fetal neurons were subjected to the scratch assay and TNF-α treatment. The neurons matured for 14 days and the scratch was initiated, for which portions of the neurons were physically scraped off the plate using a plastic cell scraper. Cells were gently washed to remove debris, and TNF-α treatment was immediately performed at a concentration of 100ng/mL. After one day, the neurites began sprouting and TNF-α treatment induced more sprouting than untreated controls. After four days, the neurites were longer and the network of neuronal processes was more complex in TNF-α treated cells when compared to untreated controls as seen in Figure 1. This experiment was repeated five times to ensure consistent results. These observations suggested that TNF-α creates a more permissive environment for neurons to extend processes into the injury site.
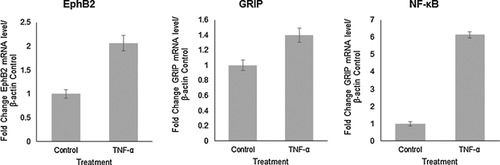
TNF-α treatment affects the expression of genes involved in the modulation of synaptic plasticity
Next, the effects of TNF-α on gene expression in a mixed human primary neuronal culture were investigated as a general outlook for genes central to synaptic plasticity and neurite remodeling. For this experiment, mixed human primary fetal neuronal cells, which can be considered as a more dynamic cell culture was used. The presence of neuronal progenitors, glia, and neurons was shown immunocytochemistry with Nestin, GFAP, and TUJ1, respectively (Fig. 2A). After one week in culture, TNF-α treatment was initiated at a concentration of 100ng/mL every four hours for three days and RNA was isolated. Following cDNA synthesis, genes central to synaptic plasticity were analyzed using a synaptic plasticity array. Several genes were upregulated upon TNF-α treatment that are directly involved in the growth and plasticity of neuronal processes, including EPHB2, GRIP1, NF-κB, KIF17, NTF3, and RELN (data not shown for KIF17, NTF3, and RELN). EphB2 was of particular interest for its role in synaptic plasticity, dendritic arborization, and axon guidance. The results were confirmed using qRT-PCR and primers specific for EphB2, GRIP, and NF-κB. Figure 2B shows that upon TNF-α treatment, EphB2 mRNA is upregulated fourfold (P < 0.00002) compared to untreated controls, and GRIP mRNA, the recruiting protein for EphB2, is upregulated threefold (P < 0.0002). NF-κB was upregulated nearly 60-fold (57-fold; P < 0.000003). The fold change was determined by first normalizing both control and TNF-α-treated samples to β-actin expression, and then the fold change for TNF-α treatment was determined by folding treated to untreated normalized values. Detected significant increase in NF-κB mRNA level suggests a possible role for NF-κB in TNF-α-induced EphB2 upregulation. The results from mRNA expression analysis in the mixed culture were confirmed in human primary fetal neurons. Indeed, the trend is the same as EphB2 is upregulated 2-fold (P < 0.0006), GRIP 1.4-fold (P < 0.004), and NF-κB 6-fold (P < 0.00001) as seen in Figure 3. Although the effects of TNF-α treatment demonstrate similar trends in isolated human primary fetal neurons, the fold changes are lower than the mixed neuronal culture, which signifies a strong involvement of glial and progenitor populations in neurite outgrowth and modulation of signaling pathway interactions in neurons. These results suggest an important role for TNF-α signaling in modulation of EphB2 expression. TNF-α has been shown to be involved in synaptic plasticity (Marchetti et al., 2004; Pickering et al., 2005; Belarbi et al., 2012; Wall et al., 2015), but a connection to EphB2 has not yet been established in human neurons.

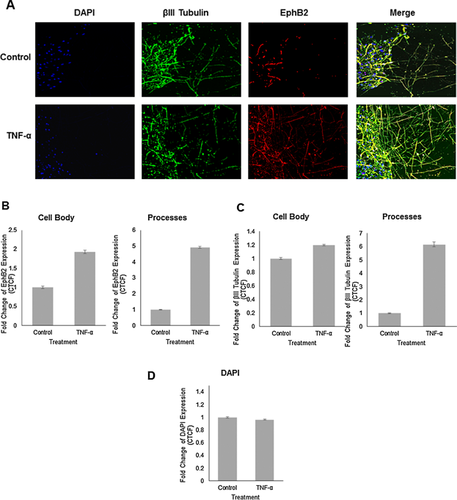
EphB2 expression is increased by TNF-α treatment in human primary fetal neurons upon induced injury
Now that the stimulatory effects of TNF-α on neurite outgrowth and expression of genes involved in synaptic plasticity modulation were established, the effect of TNF-α on EphB2 protein expression upon induced injury was investigated. As described previously, human primary fetal neurons were cultured for 14 days and the scratch injury was initiated, after which TNF-α treatment at 100ng/mL was immediately performed and repeated every 4 h for 4 days. Neurons were fixed after 4 days of treatment with TNF-α and immunolabeled with antibodies to class III β-tubulin and EphB2 (Fig. 4A). EphB2 expression in the βIII Tubulin positive cells in the cell bodies and neuronal processes that extended into the injury site was quantified (Fig. 4B). TNF-α treatment was associated with increased EphB2 expression in the cell bodies (1.8-fold, Fig. 4A) as well as in the neurites (4.8-fold, Fig. 4A) when compared to untreated controls. Incubation of human neurons with TNF-α resulted in significantly denser network of regrown neuronal processes as evident in cells stained with anti-class III β-tubulin antibody (1.2-fold increase in cell bodies and 6.2-fold increase in neuronal processes) (Fig. 4C). Fluorescence was calculated using the formula for corrected total cell fluorescence (CTCF) = Integrated Density – (Area of selected cell x Mean fluorescence of background readings) (Burgess et al., 2010). The observed EphB2 signal change is attributed to the expression of the EphB2 in elaborated processes and not to cellular proliferation as DAPI signal remains the same in control and experimental sets (Fig. 4D). In experiments described above we demonstrated that TNF-α treatment creates more permissive environment for neurons to extend processes into the injury site. This process was associated with an increase in NF-κB, EphB2 and GRIP mRNA levels. These results again confirm TNF-α mediated stimulation of regrowth of neurites after mechanical injury but also suggest that TNF-α stimulates expression of EphB2 protein in neuronal cell bodies and processes during regeneration.
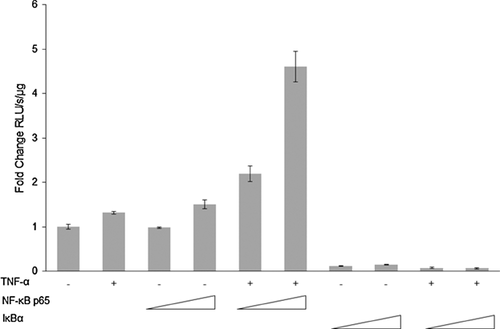
TNF-α induces EphB2 promoter activation via NF-κB signaling
In order to identify mechanisms involved in the observed increase in the levels of expression of EphB2, in silico promoter analysis with PROMO (Messeguer et al., 2002; Farré et al., 2003) was performed and three NF-κB binding sites −232 to −222 (site 1), −560 to −548 (site 2), and −730 to −719 (site 3) base pairs upstream from the transcription start site on the EphB2 promoter were identified. Since TNF-α strongly signals through NF-κB, the potential of these three binding sites for EphB2 activation was pursued. We investigated the impact of TNF-α on EphB2 promoter activity that encompasses the region starting from the position −840 to +83 base pairs using pGL3- EphB2 (−840/+83) luciferase reporter plasmid. Human primary fetal neurons were transfected using Lipofectamine 2000 with the reporter plasmid either alone or together with plasmids expressing NF-κB p65 or constitutively active IκBα mutant in the presence or absence of TNF-α treatment (100 ng/μl). As shown in Figure 5, treatment with TNF-α resulted in only a modest 1.3-fold enhancement in transcription activity driven by EphB2 promoter (−843/+83). Similar, modest dose dependent activation of EphB2 promoter activity was observed in neurons co-transfected with reporter and NF-κB expressing plasmid in the absence of TNF-α treatment (1.5 fold). The greatest induction of EphB2 promoter activity was achieved when neurons were co-transfected with reporter and NF-κB p65 expressing plasmids in combination with TNF-α treatment (100 ng/μl). A 2.2-fold to 4.6-fold induction of promoter activity was associated with increased amount of NF-κB p65 expressing plasmid used for the transfection (1 μg to 3 μg, respectively). These results led to the conclusion that while the NFκB p65 subunit can modestly activate EphB2 promoter, this ability is greatly potentiated in the presence of TNF-α. To confirm the direct involvement of p65, the p65 expression plasmid in the luciferase reporter experiment was replaced with a constitutively active IκBα mutant, which prevents translocation of p65 to the nucleus. EphB2 promoter activity was abolished even with TNF-α treatment. These results demonstrate that the presence of NF-κB p65 is essential for TNF-α-mediated activation of EphB2 promoter.
TNF-α activates EphB2 promoter via stimulation of the binding of NF-κB p65 to sites 2 and 3 on the EphB2 promoter
To determine if p65 directly interacts with the EphB2 promoter, ChIP promoter analysis was performed using human primary fetal neurons. The human fetal neurons were cultured for 14 days and either untreated or treated with TNF-α at a concentration of 100 ng/ml. DNA was prepared and incubated with 2 μg of ant-p65 antibody overnight. Control samples were incubated with IgG. The salmon sperm DNA agarose beads were then added for one hour and the DNA-antibody complex was then eluted. PCR with specific primers was performed for amplification of the DNA fragments bound to p65: NF-κB binding sites 1 (−232 to −222), 2 (−560 to −548), 3 (−730 to −719), and IκBα binding site, with latter used as a control. The EphB2 promoter schematic with binding sites is shown in Figure 6A. The distinct bands, corresponding to NF-κB binding sites 1 (−232 to −222), 2 (−560 to −548), 3 (−730 to −719), and IκBα binding site were detected in all extracts incubated with anti-p65 antibody (Fig. 6B). The intensity of the band corresponding to NF-κB binding site 2 (−560 to −548) in TNF-α treated neurons is significantly higher compared to untreated cells, suggesting that TNF-α treatment markedly enhances the binding of p65 to NF-κB binding site 2 (−560 to −548). Only modest increase in the binding of p65 to NF-κB binding site 3 (−730 to −719) was detected in the response to TNF-α treatment. Interestingly, binding of p65 to the NF-κB binding site 1 (−232 to −222) was decreased upon TNF-α treatment. DNA input was verified by extraction of DNA directly from cellular lysates (without prior immunoprecipitation) following PCR. Binding was quantified as a percent of input DNA (Fig. 6C).
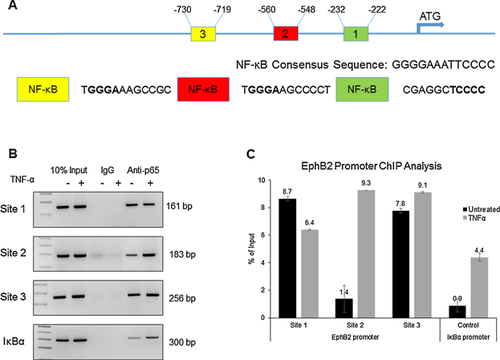
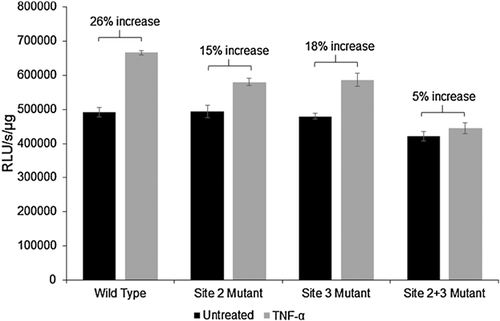
These observations point to the enhanced in vivo association of p65 with NF-κB binding site 2 (−560 to −548) in the EphB2 promoter in response to TNF-α. We hypothesize that site 2 (−560 to −548) is most responsible for TNF-α-induced activation of EphB2 signaling. To test this hypothesis, site-directed mutagenesis was utilized for our EphB2-pGL3 Basic reporter plasmid. For this, mutations were introduced in site 2 (−560 to −548) and/or 3 (−730 to −719). Site 1 (−232 to −222) was not included since there was a reduction in binding demonstrated by the ChIP assay. Human primary fetal neurons were transfected with the wild type, site 2 mutant, site 3 mutant, and combined site 2 and 3 mutant constructs after the cells were cultured for 14 days. Once the transfection was performed, TNF-α treatment at a concentration of 100ng/mL was initiated 24 h post transfection. The treatment duration was 24 h and one additional treatment was performed 30 min prior to generating lysates from the cells. Results of luciferase reporter assay show that TNF-α treatment induced a 26% increase in the activity of the wild type EphB2 promoter, a 15% increase in the activity of the EphB2 promoter with mutated NF-κB binding site 2, and 18% increase in the activity of the EphB2 promoter with mutated NF-κB binding site 3 (Fig. 7). When both sites 2 and 3 were mutated, TNF-α treatment only increased EphB2 promoter activity by 5%, confirming the results from the ChIP which demonstrated that sites 2 and 3 are primarily responsible for TNF-α-induced EphB2 promoter activation. This data provides evidence for a possible mechanism of TNF-α –mediated transcriptional regulation of EphB2.
TNF-α-induced EphB2 upregulation occurs through activation of TNFR2
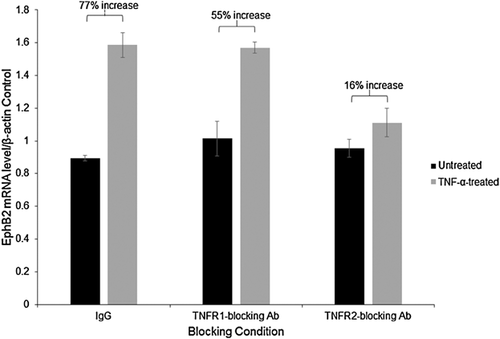
Next, we investigated TNF-α receptors involved in induction NF-κB signaling, leading to upregulation of EphB2. Human primary fetal neurons were cultured for 14 days and either TNFR1 or TNFR2 were blocked before initiating TNF-α treatment. TNFR blocking was achieved by treating the cells with either anti-TNFR1, anti-TNFR2 antibody or the nonspecific control IgG, all at a concentration of 2 μg/ml. After incubation with antibodies for 30 min, TNF-α treatment was initiated at a concentration of 100 ng/ml for 24 h, RNA was isolated and qRT-PCR was performed for EphB2 mRNA expression. EphB2 mRNA expression levels were normalized to β-actin mRNA levels. As seen in Figure 8, a 77% increase in EphB2 mRNA expression compared to untreated control was achieved by TNF-α treatment in the nonspecific control sample (Fig. 8). When TNFR1 was blocked, TNF-α induced an increase in EphB2 mRNA by 55% (Fig. 8), but only 16% when TNFR2 was blocked (Fig. 8), indicating that TNF-α induces EphB2 mRNA expression primarily through TNFR2, with a limited role of TNFR1.
Discussion
As described above, the TNF-α signaling axis and NF-κB translocation has different effects on neurons, either neuroprotective or neurotoxic. The outcome of the initiation of this signaling cascade may depend on a few factors: the cells in the CNS that are activated first, the receptor that is activated (as TNFR1 and TNFR2 have a different affinity for membrane bound TNF-α or soluble TNF-α), and the density of each receptor on the cell (Probert, 2015).
In this report we show that in human primary fetal neurons TNF-α via TNFR2 signaling enhances NF-κB binding to its consensus sequences within the EphB2 promoter and upregulates EphB2 mRNA and protein expression. This process is associated with stimulation of the growth of neuronal processes. It has been reported that many neurological adverse effects occur after anti-TNF-α therapies are given to patients with rheumatic autoimmune diseases, leading to the association with various neurological disorders, including Multiple Sclerosis (MS) (Tristano, 2010). Anti-TNF-α therapies have not been developed to specifically block signaling through either TNFR, but rather to globally block TNF-α from binding to either receptor, which has led to demyelination and various neuropathies in patients with rheumatoid arthritis (RA), Crohn's disease, and other rheumatic autoimmune diseases (Tristano, 2010).
In the in vitro “scratch” assay we observed that TNF-α treatment stimulates the regrowth of neuronal processes after mechanical injury. The process of regeneration of neurites was accompanied with an increase in NF-κB, EphB2, and GRIP mRNA levels detected by quantitative RT-PCR. The upregulation of EphB2 was of great interest because of the role of EphB2 receptors in axon guidance and dendritic arborization (Henkemeyer, 2003; Penzes et al., 2003; Murai and Pasquale, 2004; Robichaux et al., 2014). An increased expression of EphB2 protein in neuronal cell bodies and neuronal processes upon inflicted mechanical injury (“scratch”) was demonstrated by immunofluorescence. Using reporter constructs containing luciferase gene sequence placed under control of EphB2 promoter, we demonstrated that TNF-α activates the EphB2 promoter. We further investigated the possible association of NF-κB with the EphB2 promoter in the ChIP assay and showed that two NF-κB binding sites, sites 2 (−560 to −548) and 3 (−730 to −719) are primarily responsible for TNF-α-induced EphB2 promoter activation, providing further evidence for TNF-α – NF-κB mediated transcriptional regulation of EphB2. Finally, in TNFR blocking experiments, we identified TNFR2 as a main receptor involved in TNF-α signaling leading to the induction of EphB2 expression.
TNF is a ubiquitous cytokine involved in regulation of diverse biological processes including cell growth, inflammation, tumorigenesis, viral replication, infection, and autoimmunity (Aggarwal et al., 2012). TNF stimulates signaling via interaction with TNFRs. The 29 members of TNFR superfamily are divided into activating receptors and death receptors (DRs). Activating receptors, such as CD40 and TNFR2, activate NF-κB and mitogen-activated protein kinase (MAPK) pathways. DRs, such as TNFR1 and Fas, mediate extrinsic signal-induced cell death via the death domain (DD) in the intracellular region (Wu and Hymowitz, 2009), although several studies suggest that the two TNFRs can cooperate during signal transduction (Weiss et al., 1998; Mukhopadhyay et al., 2001; Aggarwal, 2003). TNF-α exerts its function via binding to two distinct transmembrane receptors: TNFR1 (also known as p55 and TNFRSF1A) and TNFR2 (also known as p75 and TNFRSF1B). TNFR1 is expressed on almost all cells of the body, but TNFR2 is expressed on certain populations of cells, including lymphocytes, endothelial cells, microglia, neurons, and oligodendrocytes (Faustman and Davis, 2013). Antagonistic functions of TNFR1 and TNFR2 also were shown during neurodegeneration in vivo with TNFR1 aggravating and TNFR2 diminishing neuronal cell loss. TNF-α can activate TNFR1 and induce neuronal apoptosis to aggregate brain tissue damage (Morganti-Kossmann et al., 2002). TNFR2 signaling was shown to have protective roles in demyelination and neuronal survival (Arnett et al., 2001; Yang et al., 2002; Marchetti et al., 2004; Chadwick et al., 2008; Rodriguez et al., 2009). TNFR2 signaling is mediated via TRAF2 and activation and nuclear entry of the pro-survival transcription factor NF-κB (Rothe et al., 1995; Carpentier et al., 2004). TNFR2 induces long term phosphatidylinositol 3-kinase-dependent NF-κB activation which is essential for neuronal survival (Marchetti et al., 2004). The inhibition of TNFR2 expression with antisense oligonucleotides was shown to sensitize neuronal-like cells toward apoptosis (Shen et al., 1997). In TNF-R2–deficient mice TNF-α was found to aggravate neuronal cell death and led to an enhancement of neurodegeneration (Fontaine et al., 2002). It appears that in TNFR2-mediated neuroprotection, the activation of NF-κB and protein kinase Akt/PKB signaling pathways plays the central role (Marchetti et al., 2004). In our studies we also observe NF-κB stimulation mediated by TNF-α -TNFR2 interaction. In addition, we found that NF-κB activation leads to increased expression of EphB2 mRNA and protein.
Cell surface Eph receptor tyrosine kinases, EphA and EphB, and their ephrin ligands regulate various aspects of cell migration, axon guidance, and synaptogenesis (Henkemeyer, 2003). Interaction of EphB receptors with ephrin-B ligands induces intracellular signals in a bidirectional manner into the receptor-expressing cell (forward signaling) and ephrin-expressing cell (reverse signaling) (Holland et al., 1996). This signaling converge on the cytoskeleton affects regulation of not only the repulsion-dependent establishment of topography and axonal growth cone collapse, but also axonal attraction (Holmberg and Frisen, 2002). Molecular mechanisms that are responsible for ephrin-induced neurite retraction involve EphB2 mediated downregulation of GTP-bound Ras (Elowe et al., 2001). Conversely, non-repulsive effects of EphB2 has been linked to the regulation of NMDA-dependent synaptic function (Grunwald et al., 2001; Henderson et al., 2001).
EphB2 upregulation by TNF-α was confirmed during the neuroregeneration observed after induced mechanical injury in the “scratch” assay, mostly in the neuronal processes extending into the scratch area. Promoter analysis with PROMO (Messeguer et al., 2002; Farré et al., 2003) in a previously published review (Pozniak et al., 2014) identified three potential NF-κB binding sites near the transcription start site of the EphB2 gene. The ChIP and site mutagenesis data shows that TNF-α signaling directly induces EphB2 in an NF-κB-dependent mechanism in human primary fetal neurons. Since the effects of TNF-α signaling are dependent on which receptor is activated, we determined that TNFR2 activation is most responsible for EphB2 upregulation with a slight increase through TNFR1. This could be due to an overlap in TNF-α signaling from both receptors as they have been shown to both activate NF-κB signaling (Probert, 2015). These data provide evidence for a neuroregenerative role of TNF-α and EphB2, as well as a novel signaling cascade for EphB2 in primary human fetal neurons. Since TNF-α levels are increased in neurodegenerative diseases, including HAND, perhaps more specific TNF-α blocking agents can be used to direct signaling through TNFR2 for beneficial signaling outcomes for neurons, including EphB2 activation. In the case of HAND, in which synaptic dysfunction and neuronal death occur, EphB2 signaling might be able to be utilized to repair damage and prevent further cognitive deficits, similarly to the results in an Alzheimer's disease model (Cissé et al., 2011).
Here we show that TNF-α–TNFR2 signaling leads to NF-κB mediated activation of EphB2 promoter which results in increased expression of EphB2 and associated enhanced growth of neuronal processes, particularly after injury. Interestingly, Schäfers and collegues demonstrated that while both TNFR1 and TNFR2 may be involved in the excitation of sensory neurons, the role of TNFR2 increases after injury (Schäfers et al., 2008). There are no reports on the possible link between TNF-α and EphB2 in neurons, although TNF-α was implicated in cooperating with the Eph/ephrin pathways in human dental pulp stem cells and bone (Zhu et al., 2015; Tejero et al., 2014). Our findings of EphB2 upregulation in TNF-α induced enhancement of neurite growth/regrowth provide insight into signaling pathways affecting regeneration of neuronal processes after injury. Further studies are warranted to decipher molecular pathways which regulate structural rearrangements in neuronal processes and the role of EphB2 upregulation in TNF-α induced enhancement of neurite growth/regrowth, which can potentially provide a therapeutic target in neuronal cell injury in the CNS.
Due to the complexity of TNF-α signaling in regards to immune activation, overlap of downstream signaling between both receptors, and variable distribution of each receptor on different cell types, EphB2 might be of greater interest as a direct therapeutic target. As demonstrated with the “scratch” assay, EphB2 appears to be driving regeneration, which would be of tremendous benefit for HAND patients and for other neurodegenerative disease patients. Although exploiting the increase in TNF-α levels in HIV for beneficial effects in the CNS would be more quickly addressed because TNF-α modulating therapies have already been developed, targeting EphB2 would not result in inflammatory effects. Whether targeting the TNF-α regulatory axis or EphB2 directly, our results demonstrate a beneficial effect of TNF-α signaling in response to induced injury and a neuroregenerative role of EphB2.
Acknowledgments
We thank past and present members of the Department of Neuroscience and Center for Neurovirology for sharing their knowledge and reagents throughout our studies. Cells and cell cultures were provided by the Basic Science Core I of the NIMH-funded Comprehensive NeuroAIDS Center (P30 MH092177). This work was presented at the 13th International Symposium on Neurovirology (San Diego, CA, June 2015). This investigation was supported in part by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award, (T32MH079785), “Interdisciplinary and Translational Research Training Program in NeuroAIDS,“ awarded to Temple University School of Medicine. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.




