Evaluation of LAMP assay using phenotypic tests and PCR for detection of blaKPC gene among clinical samples
Nianzhen Chen, Gen Li, and Yuying Si contributed equally to this work.
Funding information
This work was supported by Pudong New Area Construction of key disciplines of the Health and Family Planning Commission (grant number PW2019E-2); Shanghai Science and Technology Commission Science and Technology Innovation Action Plan (grant number 21Y11900800); the National Natural Science Foundation of China (grant number: 81971535); Shanghai Municipal Health and Planning Commission (grant number 202150010); Talent Development Program of “new star of medicine” (grant number HWRS(2020) No. 087)
Abstract
Background
Carbapenem-resistant Enterobacteriaceae (CRE) infection constitutes a public health threat, which blaKPC was the major carbapenemases concerned in China. Timely and efficient diagnosis is of paramount importance for controlling the spread of drug-resistant bacteria. Here, we develop an approach based on loop-mediated isothermal amplification (LAMP) for rapid confirmation of blaKPC within 60 min from samples collected.
Methods
We designed primers specific to detect blaKPC and evaluated it for its sensitivity and specificity of detection using real-time monitoring. Five hundred forty-six clinical specimens were analyzed by the LAMP assay and compared with the phenotypic tests and PCR. The samples with inconsistent results were further verified by Sanger sequencing.
Results
The LAMP assay displayed a detection limit of 1 × 102 CFU/ml, which was 10-fold more sensitive than the PCR. No cross-reactivity was observed for strains that produced other types of β-lactamase. Furthermore, we demonstrated concordant results (Kappa > 0.75) between the genotypic method and phenotypic tests for the 546 clinical samples. The data presented in this study suggested that the genotypic method is a reliable assay for identifying blaKPC-induced CRE in China. The results of the Sanger sequencing indicate that the developed method not only has high accuracy but also meets the need for rapid diagnosis, while the PCR method is prone to false negatives.
Conclusions
We successfully constructed a LAMP technique that can be used for auxiliary diagnosis of CRE, which is faster, cheaper, and more accurate than the PCR. It may therefore be routinely applied for detection of blaKPC producers in routine clinical laboratories.
1 INTRODUCTION
In recent years, with the widespread use of carbapenem antibiotics in clinical practice, the number of infections caused by carbapenem-resistant Enterobacteriaceae (CRE) has been increasing, and the rate of resistance of Enterobacteriaceae to carbapenem antibacterial drugs has been rapidly rising worldwide.1
Data from China Antimicrobial Surveillance Network (CHINET, www.chinets.com) in 2018 showed that the resistance rates of Klebsiella pneumoniae (Kpn) to imipenem and meropenem increased from 3.0% and 2.9% in 2005 to 25% and 26.3% in 2018, respectively. CRE has been classified as an urgent threat by the United States Centers for Disease Control and Prevention.2 Outbreak epidemics of CRE in hospitals are important factors in causing hospital-acquired infections and high patient mortality.3
The main mechanisms leading to carbapenem antibiotic resistance in Enterobacteriaceae include altered function or expression of membrane pore proteins, abnormally high expression of efflux pumps, and acquisition of enzymes capable of hydrolyzing antibacterial agents, named carbapenemases.4 The production of carbapenemases is the most common mechanism of resistance in Enterobacteriaceae, which mainly include A/B/D classes of carbapenemase. blaKPC carbapenemase, as a representative enzyme type of class A carbapenemases, was first discovered in the United States in 1998 and has been widely prevalent in many countries and regions, and is also the most common carbapenemase in clinical isolates of Kpn in China.5
Rapid detection of blaKPC genes is of great importance for clinical treatment as well as nosocomial infection prevent and control. Currently, the laboratory methods recommended by the Clinical & Laboratory Standards Institute (CLSI) for detecting carbapenemase types mainly include the modified carbapenem inactivation method (mCIM) and the Carba NP test. However, these methods are challenging to meet clinical needs due to long test cycles or cumbersome operations.6 Molecular diagnostic techniques, represented by PCR, have been widely used in pathogen resistance detection. Nevertheless, PCR assays are usually expensive and difficult to be used in less developed areas. LAMP is a new molecular detection technology that does not require high requirements for detection equipment, and can be performed using a water bath and UV lamp, which can still be effectively popularized even in less developed areas.7
Several studies have described the differences between PCR and LAMP for detecting carbapenem-resistant genes, but they have typically used clinical isolates, and few experimental studies have directly used clinical specimens for their validation and comparison.8, 9
In this study, we developed a LAMP-based method that can be used to detect the blaKPC carbapenemase gene. Meanwhile, we analyzed the results of LAMP technology indirect detection of clinical specimens in comparison with commercial PCR kits using conventional culture methods and generation sequencing technology as references.
2 MATERIALS AND METHODS
2.1 Clinical specimens and ethics statement
During the microbiological examination, sputum specimens were collected from patients suspected of lower respiratory tract infections (LRTIs) through natural expectoration or the fiberoptic bronchoscopy airway aspiration method in strict accordance with the 4th edition of the “National Clinical Inspection Operating Procedures.” Sputum samples were considered acceptable if there were >25 neutrophils and <10 squamous epithelial cells per low-power field. Midstream urine specimens were collected from patients suspected of urinary tract infections (UTI), and no antibiotics were used in the past 1 week. All clinical specimens were used in this study after performing a conventional microbiological diagnosis, and this study involved no ethical issues.
A total of 546 clinical specimens (356 sputum specimens and 190 urine specimens) were collected from patients suspected of LRTIs and UTI from September 2019 to October 2021 at Shanghai East Hospital, School of Medicine, Tongji University, which included 316 males and 230 females, aged 13–96 years, with an average age of 72.54 ± 16.50 years. In addition to infections, the vast majority of patients’ population was accompanied by other diseases (Cerebral and Cardiovascular Diseases, Cancer, Inflammation, and so on).
In our study, pathogen diagnosis was made according to the comprehensive analysis of the clinical examinations, mainly based on the results of routine bacterial culture. 356 sputum samples suspected of LRTIs detected by the conventional bacterial cultures, single infection (62.36%) accounted for the majority of the samples, and 134 samples (37.64%) were found to be a mixed infection. These positive bacterial infections primarily include Kpn (53.82%), Pseudomonas aeruginosa (22.01%), Acinetobacter baumannii (10.09%), Staphylococcus aureus (2.75%), Proteus mirabilis (2.75%), and Escherichia coli (1.53%). Urine culture was used as the confirmatory test, and there was a predominance of single infection (87.37%). The majority of isolates cultured from the 190 patients suspected of UTI were Gram-negative microorganisms with a predominance of Escherichia coli (37.40%). The next major bacteria include Kpn (28.46%), Enterococcus faecalis (13.01%), Enterococcus faecium (8.94%), Staphylococcus aureus (3.25%), Pseudomonas aeruginosa (2.44%) and Proteus mirabilis (2.44%).
2.2 Design of LAMP primers
The primers used for LAMP assay were targeted to the blaKPC genes were obtained from NCBI databases (https://www.ncbi.nlm.nih.gov/pathogens/) and were designed using the Primer Explorer V5 software (Eiken Chemical Co., Ltd., Tokyo, Japan; http://primerexplorer.jp/lampv5e/index.html). These primers include two outer primers (F3 and B3), two inner primers (FIP and BIP), and loop primers (LF, LB). The primers shown in Table 1 were synthesized by Invitrogen (Carlsbad, CA, USA). The primers were formulated into a solution containing 0.2 µM of outer primers, 1.6 µM of inner primers, and 0.6 µM of loop primers, and stored at −20°C.
| Primer | Sequence (5′→3′) |
|---|---|
| blaKPC-F3 | GGCTCAGGCGCAACTG |
| blaKPC-B3 | GGGTGACCACGGAACCA |
| blaKPC-FIP | CGGCAGCAAGAAAGCCCTTGAATTTTTAAGTTACCGCGCTGAGGA |
| blaKPC-BIP | TGTGCTGGCTCGCAGCCATTTTGCGCATTTTTGCCGTAACGG |
| blaKPC-LB | GGCGCAACTGTAAGTTACCG |
2.3 The LAMP assay was established and optimized
Detailed information of the reaction system has been published previously by our laboratory.10 The LAMP reaction was carried out in a 20-µl volume reaction mixture containing the following reagents: 2.5 µl 10× ThermoPol Reaction Buffer, 1 µl Bst 2.0 DNA polymerase [both New England Biolabs (Beijing) Ltd., Beijing, China], 9.5 µl mixture (with ddH2O, Mg2+), 1 µl SYBR‑Green I (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), 1 µl primers (0.2 µM of the outer primer and 1.6 µM of the inner primer), and 5 µl template. The LAMP assay was optimized at 63°C for 45 min. In addition, the optimal amplification concentration of Mg2+ was determined to be 8 mM.
2.4 Bacterial isolates
Nineteen whole-genome sequencing-verified "standard strains" producing other representative β-lactamases from the Microbiology Laboratory of Shanghai East Hospital, School of Medicine, Tongji University, were collected (Table 2). Bacterial strains were grown on Columbia sheep blood agar (Thermo Scientific, KS, USA) without antibiotics overnight at 37℃ fresh overnight bacterial cultures were used for experimental studies.
| Strain no. | Strain species | β-Lactamase genes | Enzyme family (Ambler classification) |
|---|---|---|---|
| 1 | Klebsiella pneumoniae | bla KPC-2 | Carbapenemase (A) |
| 2 | Klebsiella oxytoca | bla KPC-2 | Carbapenemase (A) |
| 3 | Pseudomonas aeruginosa | bla KPC-2 | Carbapenemase (A) |
| 4 | Klebsiella pneumoniae | bla IMP-4 | Carbapenemase (B) |
| 5 | Enterobacter cloacae | bla NDM-1 | Carbapenemase (B) |
| 6 | Escherichia coli | bla NDM-1 | Carbapenemase (B) |
| 7 | Pseudomonas aeruginosa | bla VIM-2 | Carbapenemase (B) |
| 8 | Acinetobacter baumannii | bla OXA-23 | Carbapenemase (D) |
| 9 | Acinetobacter baumannii | bla OXA-24 | Carbapenemase (D) |
| 10 | Klebsiella pneumoniae | bla OXA-48 | Carbapenemase (D) |
| 11 | Escherichia coli | bla CTX-3 | Extended spectrum β-lactamases (A) |
| 12 | Klebsiella pneumoniae | bla CTX-15 | Extended spectrum β-lactamases (A) |
| 13 | Escherichia coli | bla CTX-55 | Extended spectrum β-lactamases (A) |
| 14 | Escherichia coli | bla CTX-64 | Extended spectrum β-lactamases (A) |
| 15 | Escherichia coli | bla CTX-14 | Extended spectrum β-lactamases (A) |
| 16 | Klebsiella pneumoniae | bla SHV-1 | Penicillinase (A) |
| 17 | Escherichia coli | bla TEM-1 | Penicillinase (A) |
| 18 | Escherichia coli | bla DHA-1 | Cephalosporinase (C) |
| 19 | Morganella morganii | bla CMY-2 | Cephalosporinase (C) |
2.5 Evaluation of the specificity and sensitivity of the LAMP assay
The standard bacterial strains Kpn ATCC strain BAA1705 were used in assay validation as positive control for blaKPC. It was purchased from Shanghai Beinuo Biotechnology Co., Ltd. (Shanghai, China).
To assess the specificity of the developed LAMP system, nucleic acids from Kpn ATCC strain BAA1705 were used to positive control for blaKPC, and ddH2O was used to be negative control. Nucleic acids from other β-lactamases strains (Table 2) were used to test the developed system to determine the cross-reactivity with the blaKPC gene. All experiments were repeated three times.
To evaluate the sensitivity of the developed LAMP system, the original BAA1705 bacterial suspension as described above was ten-fold serially diluted, ranging from 107 to 101 CFU/ml,11 and each dilution was used for the system as described above to test the sensitivity. Every experiment was repeated three times.
2.6 Evaluation of the LAMP assay using clinical samples
For the 546 clinical specimens (356 sputum specimens and 190 urine specimens), a parallel study using both the SCM, PCR, and LAMP assays was carried out. To extract DNA from the sputum specimens, an equal volume of 4% NaOH was added to liquefy the sample, placed on a vortex shaker to disperse the specimens as much as possible, and then incubated at 37°C for 30 min. If the specimen was viscous, the volume of NaOH was increased, or the liquefaction time extended as appropriate. The sputum specimens have to be free of viscosity after liquefaction. To extract DNA from the urine specimens, 2 ml sample was centrifuged at 12,000 g for 10 min, and resuspended in 500 ul PBS (pH 8.0).
Total genomic DNA was extracted from the liquefied sample, using a nucleic acid releasing agent extraction kit (Fosun Pharmaceutical Co., Ltd., Shanghai, China), according to the manufacturer's instructions. DNA was frozen at −80°C in 5-µl aliquots and then tested with the LAMP system we established. ddH2O was used as negative control, and the standard strain was used as the positive control. All the LAMP assays were performed on the ABI 7500 (Applied Biosystems, Foster City, CA, USA) and analyzed using ABI 7500 Software Version 2.3.
2.7 Results were compared between the LAMP and other methods
According to the manufacturer's instructions, all isolates were identified using the MALDI-TOF Biotyper system (Bruker Daltonics, Bremen, Germany) with the “formic acid extraction” procedure. The accuracy of the identification results was assessed by the Log Score values obtained from the MALDI-Biotyper software, ranging from 0 to 3.0. In addition, the strains in this experiment were Enterobacterales bacteria with Log Score values greater than 2.0. Antimicrobial susceptibility testing (AST) was performed in an automated manner by the VITEK-2 Compact system (BioMerieux, Marcy l'Etoile, France), and minimal inhibitory concentration (MIC) of carbapenems was reconfirmed using the E-test (Kont Biology Technology, Wenzhou, China). The results of AST were interpreted according to CLSI.12 All CRE strains were resistant to at least one carbapenem (ertapenem-MIC ≥ 2 μg/ml, imipenem-MIC ≥ 4 μg/ml, meropenem-MIC ≥ 4 μg/ml). It is the phenotypic tests carried in this study.
Routine screening for blaKPC was performed via PCR detection using the Carbapenem Resistance Gene blaKPC Detection Kit (Fluorescence PCR) (Shanghai Zhijiang Biotechnology Co. Ltd., China). The experimental operation was carried out according to the kit operating instructions.
2.8 DNA sequencing
To analyze all the inconsistent samples between the LAMP assay and other methods, 20 µl of sample DNA was sent to Sangon Biotech Co., Ltd. (Shanghai, China) for Sanger sequencing. Designed Sanger sequencing primers for blaKPC were as follows: forward primer, CCGACGCCTTGCCAATTGCAGA; reverse primer, CCGCCGCCAATTTGTTGCT. The products are electrophoresed on a PAGE gel. The purified products were then subjected to Sanger sequencing using a BigDye Terminator v1.1 and ABI 3730XL DNA Analyzer (ABI, Carlsbad, CA, USA).
2.9 Data processing and analysis
SPSS statistical software version 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 7 software (GraphPad Prism Software, La Jolla, CA, USA) were used for statistical analysis and figures. All the 546 samples were detected by the LAMP and other methods. The agreement between these two methods was evaluated by κ coefficient with a 95% CI for each pathogen, and κ ≥ 0.75 indicates excellent agreement, 0.75 > κ ≥ 0.4 indicates fair to moderate agreement, and κ < 0.4 indicates poor agreement. Sensitivity, specificity, positive and negative predictive values, positive and negative likelihood ratios were also analyzed by SPSS.
3 RESULTS
3.1 Specificity of the LAMP assay
The specificity of primers is critical for accurate detection of the LAMP assay. We assessed the specificity of the LAMP assay for detecting blaKPC producers by evaluating its reactivity with strains that produced other types of β-lactamase (N = 16). Kpn ATCC strain BAA-1705 was used as the positive control and double-distilled water as the negative control. As shown in Figure 1, when the corresponding strains’ DNA was added, only the DNA from blaKPC producers (isolated bacterial strains No.1, 2, 3, and positive control) had an amplification curve, and other types of β-lactamase and the negative control showed no signal. Briefly, no cross-amplification was observed in the LAMP, indicating the assay was highly specific for detecting blaKPC.
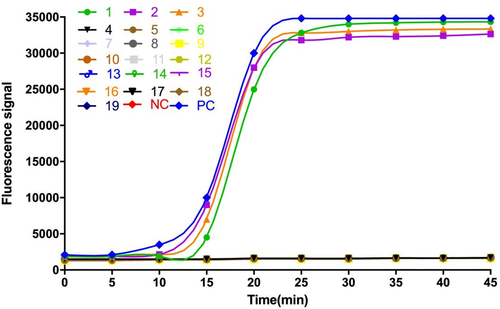
3.2 Sensitivity test for the LAMP assay
To evaluate the detection limit of the LAMP assay, DNA extracted from cultured Kpn ATCC BAA-1705 cells was ten-fold serially diluted, ranging from 1 × 107 CFU to 1 × 101 CFU/ml. The results were also compared with those of a commercial PCR kit. As illustrated in Figure 2, the detection limit of the LAMP was even from 102 CFU/ml, whereas the PCR was only able to amplify 103 CFU. Thus, the sensitivity of LAMP was higher than the PCR, and all amplification curves appeared within 30 min.
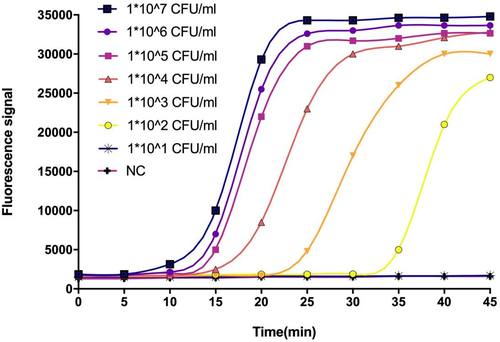
3.3 Clinical performance of the developed LAMP assay
To illustrate the performance of our established method, 546 clinical specimens (356 sputum samples and 190 urine samples) were tested simultaneously using the LAMP assay, the commercial PCR kit, and the phenotypic tests with conventional bacterial cultures. The results are presented in Table 3. It has been shown that strains producing blaKPC predominate among CRE in China. Therefore, we classified the phenotypic test results of 546 specimens into CRE and non-CRE, including 308 CRE (252 sputum cases and 56 urine cases) and 238 non-CRE (104 sputum cases and 134 urine cases). Results of the LAMP assay and a commercial PCR kit, respectively, were compared with results using phenotypic tests. Of 308 CRE samples, 292 (240 sputum cases and 52 urine cases) were detected as positive by the commercial PCR kit, and 297 (245 sputum cases and 52 urine cases) were tested as positive by the LAMP assay. Sanger sequencing was further used to analyze the inconsistent samples among the three methods. We observed four CRE sputum cases with positive LAMP results but negative PCR results. Five were confirmed as true-positive results (Table 4). With the Sanger sequencing, a fraction of CRE was confirmed not to produce blaKPC carbapenemase. We compared the results from genotypic and phenotypic tests, and Kappa statistical analysis in urine indicated no difference between the commercial PCR kit and the LAMP assay. However, of the 104 non-CRE sputum specimens, 14 (13.5%) cases with positive PCR results but 24 (23.1%) cases with positive LAMP results. Better agreement was observed between the commercial PCR kit and the phenotypic tests. Of the samples that were non-CRE by the phenotypic tests, 42 were carbapenem-resistant Acinetobacter baumannii (CRAB) or carbapenem-resistant Pseudomonas aeruginosa (CRPA) by the phenotypic tests. Among the 42 clinical samples, 11 were detected as positive by the LAMP assay, while only 5 were tested positive by the commercial PCR kit. As can be seen from Table 3, 9 samples were confirmed positive by amplicon sequencing of the blaKPC gene. Whether CRE or CRAB, or CRPA, we note that the rate of false-negative detection of the commercial PCR kit is higher than the developed LAMP assay. In addition, 19 cases demonstrated susceptibility to carbapenems with blaKPC by the LAMP assay. Seventeen cases were confirmed positive by Sanger sequencing, but the commercial PCR kit detected only 11 cases. Briefly, the sensitivity of the commercial PCR kit was lower than the LAMP when the sequencing results were used as control, resulting in some positive specimens being missed.
| Sample types | Phenotypic tests | RT-PCR | LAMP | Subtotal | Kappa | |||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | RT-PCR/phenotypic tests | LAMP/phenotypic tests | |||
| Sputum | CRE | 240 (95.2%)a | 12 (4.8%)c | 245 (97.2%)a | 7 (2.8%) c | 252 | 0.822 | 0.779 |
| non-CRE | 14 (13.5%)b | 90 (86.5%)d | 24 (23.1%)b | 80 (76.9%)d | 104 | |||
| Subtotal | 254 | 102 | 269 | 87 | 356 | |||
| Urine | CRE | 52 (92.9%)a | 4 (7.1%)c | 52 (92.9%)a | 4 (7.1%) c | 56 | 0.875 | 0.875 |
| non-CRE | 6 (4.5%)b | 128 (95.5%)d | 6 (4.5%)b | 128 (95.5%)d | 134 | |||
| Subtotal | 58 | 132 | 58 | 132 | 190 | |||
- a The rate of sensitivity or true-positive rate.
- b False positive rate.
- c False-negative rate.
- d The rate of specificity or true negative rate.
| Phenotypic tests | RT-PCR/LAMP | Sanger sequencing | Total | |
|---|---|---|---|---|
| + | − | |||
| CRE | +/− | 0 | 1 | 1 |
| −/+ | 5 | 2 | 7 | |
| −/− | 0 | 8 | 8 | |
| CRAB & CRPA | +/+ | 5 | 0 | 5 |
| −/+ | 4 | 2 | 6 | |
| CS | +/+ | 11 | 0 | 11 |
| −/+ | 6 | 2 | 8 | |
- Abbreviations: CRAB & CRPA, carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Pseudomonas aeruginosa; CS, carbapenem sensitive.
4 DISCUSSION
Carbapenems were widely used as effective drugs in treating multidrug-resistant bacterial infections. The treatment options for drug-resistant Gram-negative bacteria, such as CRE and CRAB, are limited. Multi-drug-resistant organisms such as CRE have become a challenge to patients, clinicians, and public health.13 CRE has been classified as an urgent threat, and it is urgent to prevent the outbreaks of its. However, conventional identification and susceptibility testing methods of microorganisms usually require at least 2 days from specimen collection. The time required for completing the whole process is long, and it is prone to false negatives.
Currently, clinical laboratory testing for CRE included the mCIM tests and further modification to mCIM with the addition of EDTA (eCIM) synergy test, the modified Hodge test (MHT), and the Carba NP test. The MHT is the first recommended method by CLSI growth-based carbapenemase detection test for isolated strain in 2009 with a high level of sensitivity and specificity.14, 15 The advantages of the MHT are cheap and easy to perform. However, it is difficult to interpret some results, distinguish enzyme types, and be time-consuming. So, it was removed from the CLSI M100-S28 document in 2018. The Carba NP test was performed on CRE isolates according to the CLSI M100-S25 to detect the presence of carbapenemase in 2015. However, it is cumbersome to operate and requires special reagents. It is not suitable for routine laboratory work. The mCIM combined with eCIM was recommended according to CLSI 2018 standards, distinguishing between serine and metallo-carbapenemases.16 The phenotypic detection, mCIM combined with eCIM, showed high sensitivity and specificity to detect carbapenemase-producing Enterobacteriaceae (CPE), compared with the MHT.17 Lately, various novel methods for carbapenemases detection have been proposed, such as flow cytometry or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS).18, 19 However, neither of them had a standardized procedure. In addition, they are still too expensive for a wide range of clinical applications. All methods mentioned above are performed from isolated strains, which are not helpful in slowing down the spread of CRE in time.
Thus, to meet the challenge, this study developed a LAMP-based method to detect CRE with high sensibility and specificity based on the actual situations in China, which will benefit clinical laboratories with the earlier detection of CRE.
In the present study, to determine the clinical performance of the LAMP assay, a total of 546 clinical samples (356 sputum samples and 190 urine samples) were screened by LAMP, PCR, and AST simultaneously. Applying culture results (phenotypic results) as the gold standards, there was no difference between the developed LAMP assay and the commercial PCR kit in 190 urine samples. More than 0.85 revealed an excellent agreement between the genotypic methods and phenotypic tests. Four CRE cases were identified by the phenotypic tests but tested negative by the developed LAMP assay and the commercial PCR kit. We considered it is possible to produce other carbapenemases and were further confirmed by Sanger sequencing. In addition, six urine samples blaKPC were detected by genotypic methods, but phenotypic tests suggested susceptibility to carbapenems. Of these, 4 patients had a previous history of CRE infection. We believed that the difference might have resulted from samples. CRE was not the dominant microflora responsible for this UTI, so it was not detected in conventional bacterial culture methods. Altogether, we conclude that the results between the developed LAMP assay and the commercial PCR kit are consistent and in excellent agreement with the phenotypic results. The LAMP assay facilitates the timely detection and control of the source of CRE infection.
Gram-negative bacteria, especially Enterobacteriaceae, are the leading cause of LRTIs. Rapid screening to diagnose CRE infection is also essential for controlling the nosocomial infection. Of 356 sputum samples collected from patients were signs and symptoms of LRTIs, there were 252 identified as CRE by the phenotypic tests, and only 240 samples were detected by the commercial PCR kit, but the LAMP detected 245. There were 4 CRE cases caused by other carbapenemases and verified by Sanger sequencing. Also, a proportion of the non-CRE specimens with CRAB and CRPA were not detected by the commercial PCR kit. However, positive results by the developed LAMP, which was validated by Sanger sequencing further, are false-negative results in the commercial PCR kit. The LAMP reaction is not susceptible to the influence of the different components often present in clinical samples.20 Thus, it is not necessary to purify DNA from samples with higher sensitivity. While analyzing, we noticed the presence of a proportion of susceptibility to carbapenems in sputum samples that were detected blaKPC. This situation occurred in sputum samples more frequently than urine samples. This may be due to heterogeneous resistance of bacteria leading to false-negative results in routine drug sensitivity tests, or it may be that there are dead or low-level blaKPC expressing pathogens in the test specimen that LAMP can detect, but routine tests cannot.
A few studies have evaluated CRE detection by genotypic tests, for instance, PCR, LAMP, and high-throughput next-generation sequencing.21 To our knowledge, however, none of these studies used clinical samples directly but isolated strains after bacterial culture. Plasmids encoding carbapenemases have been demonstrated to play a core role in the rapid spread of CRE,22 among which blaKPC and NDM were the major carbapenemases concerned in China.23 We developed a method based on the actual situations in China. The data presented in this study suggested that the developed LAMP assay is better than the commercial PCR kit. What is more, it must be pointed out that the LAMP can not only have high accuracy but also meet the need of rapid diagnosis, which can be completed within 60 min from specimen collection. Thus, this method can be helpful in a large-scale survey of blaKPC and controlling the spread of blaKPC-induced CRE in this region. Nevertheless, limitations to this study primarily are that only one type of carbapenemases was tested in this study. Future studies are needed to confirm other carbapenemases, including blaNDM, blaOXA-23, blaOXA-48, and so on.
In conclusion, we propose a new method for auxiliary diagnosis of CRE and with lower cost. It has a positive significance in the early treatment of CRE. These results indicate that the developed method has good potential for application in clinical.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.




