Development of a novel five-lncRNA prognostic signature for predicting overall survival in elderly patients with breast cancer
Yang Luo and Yue Zhang authors contributed equally to this work.
Funding information
This study was supported by the National Natural Science Foundation of China (81972016), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (Grant No.2017-I2 M-3-012 & No.2017-I2 M-1-013), and Beijing Municipal Natural Science Foundation (7162164).
Abstract
Background
Breast cancer (BC) is an age-related disease. Long noncoding RNAs (lncRNAs) have been proven to be crucial contributors in tumorigenesis. This study aims to develop a novel lncRNA-based signature to predict elderly BC patients’ prognosis.
Methods
The RNA expression profiles and corresponding clinical information of 182 elderly BC patients were retrieved from The Cancer Genome Atlas (TCGA). Differentially expressed lncRNAs (DElncRNAs) between BC and adjacent normal samples were used to construct the signature in the training set through univariate Cox regression analysis, LASSO regression analysis, and multivariate Cox regression analysis. Kaplan–Meier analysis and time-dependent receiver operating characteristic (ROC) analysis were used to evaluate the predictive performance. Besides, we developed the nomogram. Gene set enrichment analysis (GSEA) was performed to reveal the underlying molecular mechanisms.
Results
We constructed the five-lncRNA signature (including LEF1-AS1, MEF2C-AS1, ST8SIA6-AS1, LINC01224, and LINC02408) in the training set, which successfully divided the patients into low- and high-risk groups with significantly different prognosis (p = 0.000049), and the AUC at 3 and 5 years of the signature was 0.779 and 0.788, respectively. The predictive performance of this signature was validated in the test and entire set. The 5-lncRNA signature was an independent prognostic factor of OS (p = 0.007) and the nomogram constructed by independent prognostic factors was an accurate predictor of predicting overall survival probability. Besides, several pathways associated with tumorigenesis have been identified by GSEA.
Conclusions
The 5-lncRNA signature and nomogram are reliable in predicting elderly BC patients’ prognosis and provide clues for clinical decision-making.
1 INTRODUCTION
Breast cancer (BC) is the most common type of cancer and one of the leading causes of cancer death among females worldwide. The incidence of BC increases with age, with a risk of 0.5% for women aged 30, 2.3% for women aged 50, and 3.9% for women aged 70.1 At present, around 30%–35% of BC patients are above 70 years of age at onset of diagnosis, and this is expected to change dramatically in the years ahead as a result of increasing life expectancy and an overall aging population.2 Despite this, the currently available literature shows a remarkable lack of older women in clinical trials. Providing high-level care for an increasing number of older BC patients is of critical importance.
When compared with younger patients, BCs in older women appear to develop relatively more “indolent” tumors characterized by higher endocrine receptor expression, lower proliferative indices, and less likely to have overexpression of HER2.3 Despite the fact that older patients with BC are more likely to have favorable tumor characteristics, outcomes do not reflect this apparent advantage. Instead, BC-specific survival is less for older patients when adjusted for age, suggesting that these patients are not being given the state-of-the-art treatment, which potentially increases their risk of disease recurrence and mortality. On the other hand, older women with BC show a high prevalence of comorbidities that result in a variable decline in organ function that may compromise their BC care. Although most BCs in older women are identified at an early, treatable stage, treatment decisions must consider the reduction in recurrence risk that would be gained by specific therapies and balance that risk with the potential for treatment-related toxicity.4 Because recurrent disease remains a persistent and vexing problem, deciding which patients are optimal candidates for adjuvant treatment in light of the potent toxicities as well as potential benefits of adjuvant treatment is challenging.5 Therefore, it is an urgent clinical need to identify those who are more biologically aggressive and tend to recur early.
With the development of high-throughput sequencing technology, long noncoding RNAs (lncRNAs) have attracted increasing attention. LncRNAs are a class of non-protein coding transcripts with length longer than 200 nucleotides that can play crucial regulatory roles in tumorigenesis by regulating gene expression at epigenetic, transcriptional, and post-transcriptional levels.6 Emerging evidence suggested that lncRNAs might serve as prognostic biomarkers in several tumor types to evaluate patients’ prognosis. Sun et al. reported that a six-lncRNA signature (SACS-AS1, MME-AS1, CSMD2-AS1, RP11-360F5.1, RP11-25K19.1, and CTC-467M3.1) was detected as an independent prognostic factor for diffuse large-B-cell lymphoma patients.7 In another study, the researchers confirmed that a four-lncRNA signature ((TCL6, PVT1, MIR155HG, and HAR1B) had significant prognostic value in clear cell renal cell carcinoma.8 Some studies reported that lncRNA-based signatures could be used as prognostic biomarkers in BC.9-11 However, none of the researches focused on elderly BC patients.
In this study, the age threshold of elderly patients was set as 70 years. We analyzed sequencing data from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) and constructed a novel lncRNA-based signature for overall survival (OS) prediction in elderly BC patients. Based on the prognostic assessment of our signature, we can develop individualized treatment strategies for each patient to improve treatment effectiveness and long-term survival. Besides, gene set enrichment analysis (GSEA) revealed significant activation of biological functions in high-risk patients, providing a new perspective for the research of elderly BC patients. Moreover, the prognosis of elderly BC patients can be evaluated more effectively through the nomogram based on our signature and clinical characteristics.
2 MATERIAL AND METHODS
2.1 Data collection and preprocessing
The RNA expression profiles and corresponding clinical information of 182 elderly BC patients were retrieved from TCGA in March 2020. All enrolled patients met the following inclusion criteria: (1) pathological diagnosis was BC; (2) females greater than or equal to 70 years of age; (3) the follow-up time was at least 30 days; (4) no treatment was received before being enrolled in TCGA; (5) not synchronized with other malignancy. Since the data were derived from the TCGA project, the approval of institutional ethics committees was not needed.
The RNA sequencing data of enrolled patients included 182 BC samples and 18 adjacent normal samples. LncRNAs and mRNAs were annotated by the human gene annotation file (GRCh38.p13) obtained from the Ensembl project (http://asia.ensembl.org/index.html). Considering that some lncRNAs with little or no expression in some tissues are not very important, only those with raw count value ≥3 in more than 75% of samples were retained. The edgeR package12 in R was used to identify the differentially expressed lncRNAs (DElncRNAs) between BC and adjacent normal samples with the cutoff of |log2 fold change (log2FC) | >2 and adjusted p-value < 0.05. The normalized expression level of each DElncRNA was log2-transformed for the following processing.
2.2 Construction and validation of a lncRNA-based prognostic signature
All elderly BC patients were randomly divided into a training set (N = 91) and a test set (N = 91). In the training set, we conducted a univariate Cox proportional hazards regression analysis to screen out the DElncRNAs significantly associated with patients’ OS. LncRNAs with a p-value of less than 0.1 were considered to be significantly correlated with prognosis and entered into subsequent processing. After that, we applied the least absolute shrinkage and selector operation (LASSO) regression to reduce the variables where the λ value was determined by 10-fold cross-validation.13 Then, the multivariate Cox proportional hazards regression analysis was performed to construct the optimal lncRNA-based prognostic signature. The lncRNA-based risk score formula was built as following: Risk Score = β1 ∗ exp1 + β2 ∗ exp2 + · · · + βn ∗ expn. In this formula, β represents the coefficient of lncRNA in the result of multivariate Cox regression analysis, and exp means normalized and log2-transformed expression level of lncRNA.
Patients in the training set were divided into a low-risk group and a high-risk group according to the median risk score. The differences of OS between the two groups were analyzed using the Kaplan–Meier method and compared by the log-rank test. The time-dependent receiver operating characteristic (ROC) curve was employed to assess the predictive ability of the lncRNA-based prognostic signature. Meanwhile, we validated this signature in the test set and the entire set.
2.3 Prognostic assessment of the prognostic lncRNA signature
The predictive performance of the lncRNA-based prognostic signature was further assessed in the entire set. We performed the univariate and multivariate Cox regression analyses to verify whether the lncRNA-based signature is an independent prognostic factor of OS for elderly BC patients. Then, the risk stratification analysis was used to test whether our signature could effectively predict within the same clinical factor. Furthermore, the time-dependent ROC analysis was conducted to compare the lncRNA-based signature's predictive accuracy with other clinical characteristics.
2.4 Development of a prognostic nomogram
To provide clinicians with a quantitative tool for assessing the probability of patients’ OS, we developed a nomogram in combination with the lncRNA-based signature and clinical independent prognostic factors. Calibration plots were used to evaluate the agreement between predicted and actual survival probabilities. The 45-degree line represented the perfect prediction model. The actual probability of OS and predicted results of the nomogram were plotted on the x-axis and y-axis, respectively. Moreover, the discriminative ability of the prognostic nomogram was measured by concordance-index (C-index).
2.5 Gene set enrichment analysis
According to the cutoff obtained from the training set, all elderly BC patients were categorized into low- and high-risk groups. Gene set enrichment analysis (GSEA) was performed by the GSEA software (v.4.0.3)14 to investigate the significantly altered biological states or processes between the two groups. The hallmark gene sets (h.all.v7.1.symbols.gmt)15 and the KEGG pathway gene sets (c2.cp.kegg.v7.1.symbols.gmt), downloaded from the Molecular Signatures Database (MSigDB), were chosen as the reference gene sets. Normalized enrichment score (NES) >1.5 and false discovery rate (FDR) value <0.05 after performing 1000 random permutations were defined as the cutoff criteria.
2.6 Statistical analysis
All statistical analyses were performed using R software version 3.6.1 and SPSS version 22.0. The Mann–Whitney U test and Fisher's exact test were used to compare the differences between continuous and categorical variables between the training set and test set, respectively. The heatmaps were generated via an R package of pheatmap. The survival package was applied to conduct the Kaplan–Meier survival analysis, univariate Cox regression analysis, and multivariate Cox regression analysis. The glmnet package was used to perform LASSO regression. The time-dependent ROC curve was depicted by an R package called “timeROC”. Furthermore, the prognostic nomogram, calibration plots, and C-index were performed using the rms package. A p-value of < 0.05 was considered statistically significant.
3 RESULTS
3.1 Patient characteristics
A total of 182 elderly BC patients were included in our study. The clinical characteristics of the training set and test set were listed in Table 1, including age, race, pathological type, stage, ER status, PR status, and HER2 status. There was no significant difference in clinical characteristics between the two data sets (all p > 0.05).
| Characteristics | Training set (N = 91) | Test set (N = 91) | p-Value |
|---|---|---|---|
| Age (years) | 76 (73,79) | 76(73,80) | 0.283 |
| Race | 0.712 | ||
| Asian | 1 (1.1%) | 3 (3.3%) | |
| Black or African American | 17 (18.7%) | 14 (15.4%) | |
| White | 64 (70.3%) | 63 (69.2%) | |
| Unknown | 9 (9.9%) | 11 (12.1%) | |
| Pathological type | 1.000 | ||
| Infiltrating duct carcinoma | 54 (59.3%) | 54 (59.3%) | |
| Lobular carcinoma | 21 (23.1%) | 21 (23.1%) | |
| Others | 16 (17.6%) | 16 (17.6%) | |
| T stage | 0.276 | ||
| T1 | 29 (31.9%) | 25 (27.5%) | |
| T2 | 40 (44.0%) | 52 (57.1%) | |
| T3 | 16 (17.6%) | 9 (9.9%) | |
| T4 | 6 (6.6%) | 5 (5.5%) | |
| N stage | 0.741 | ||
| N0 | 49 (53.8%) | 43 (47.3%) | |
| N1 | 21 (23.1%) | 28 (30.8%) | |
| N2 | 11 (12.1%) | 9 (9.9%) | |
| N3 | 7 (7.7%) | 9 (9.9%) | |
| Unknown | 3 (3.3%) | 2 (2.2%) | |
| M stage | 0.794 | ||
| M0 | 70 (76.9%) | 67 (73.6%) | |
| M1 | 1 (1.1%) | 2 (2.2%) | |
| Unknown | 20 (22.0%) | 22 (24.2%) | |
| Tumor stage | 0.565 | ||
| I | 23 (25.3%) | 16 (17.6%) | |
| II | 41 (45.1%) | 50 (54.9%) | |
| III | 25 (27.5%) | 21 (23.1%) | |
| IV | 1 (1.1%) | 2 (2.2%) | |
| Unknown | 1 (1.1%) | 2 (2.2%) | |
| ER status | 0.496 | ||
| Negative | 19 (20.9%) | 14 (15.4%) | |
| Positive | 67 (73.6%) | 74 (81.3%) | |
| Unknown | 5 (5.5%) | 3 (3.3%) | |
| PR status | 0.459 | ||
| Negative | 28 (30.8%) | 24 (26.4%) | |
| Positive | 57 (62.6%) | 64 (70.3%) | |
| Unknown | 6 (6.6%) | 3 (3.3%) | |
| HER2 status | 0.298 | ||
| Negative | 45 (49.5%) | 49 (53.8%) | |
| Positive | 13 (14.3%) | 18 (19.8%) | |
| Unknown | 33 (36.3%) | 24 (26.4%) | |
- Abbreviations: ER, estrogen receptor; HER2, human epithelial growth factor receptor 2; M, distant metastasis; N: lymph node status; PR, progesterone receptor; T, tumor size.
3.2 Identification of DElncRNAs in elderly BC patients
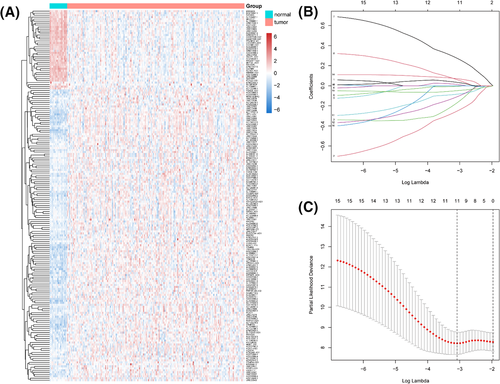
Following the cutoff criteria: |log2FC| >2 and adjusted p-value < 0.05, a total of 187 DElncRNAs were identified between 182 BC samples and 18 adjacent normal samples, including 148 upregulated lncRNAs and 39 downregulated lncRNAs. The heatmap of DElncRNAs was shown in Figure 1A. These lncRNAs were retained as candidate prognostic biomarkers for the following data processing.
3.3 Construction of the 5-lncRNA prognostic signature in the training set
First, the univariate Cox regression analysis found out that 15 lncRNAs (AC021087.4, AC099850.4, AGAP1-IT1, AP005131.3, CADM3-AS1, LEF1-AS1, LINC01136, LINC01224, LINC01238, LINC02408, MEF2C-AS1, RHPN1-AS1, SCGB1B2P, ST8SIA6-AS1, and WDR86-AS1) were significantly correlated with the overall survival of elderly BC patients (all p < 0.1). Then, 11 robust lncRNAs (AC021087.4, AGAP1-IT1, AP005131.3, LEF1-AS1, LINC01136, LINC01224, LINC01238, LINC02408, MEF2C-AS1, SCGB1B2P, and ST8SIA6-AS1) were screened out by using the LASSO regression in these 15 candidate lncRNAs (Figure 1B, C). Finally, we conducted the multivariate Cox regression analysis to construct a lncRNA-based prognostic signature based on the expression level and coefficient of five lncRNAs as follows: Risk score = (−0.735918381* LEF1-AS1) + (−0.306357639* LINC01224) + (0.636798285* LINC02408) + (−0.482279544* MEF2C-AS1) + (0.154054862* ST8SIA6-AS1). In this signature, MEFC-AS1 was significantly downregulated in the BC tissues, while the other four lncRNAs were upregulated considerably.
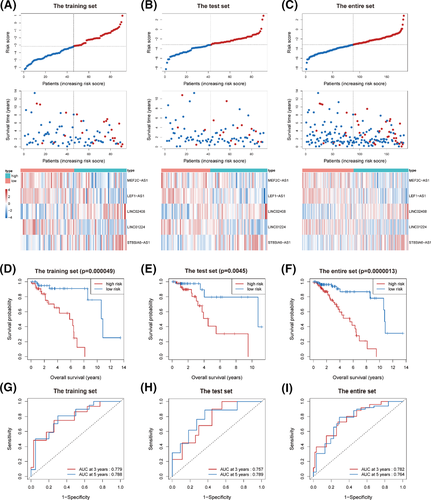
According to the prognostic model formula, we calculated the patients’ risk score in the training set and assigned them to the low-risk group and the high-risk group using the median risk score (−3.161) as the threshold. The distribution of risk score and patient survival status was shown in Figure 2A, and indicated that the patients in the high-risk group had shorter survival time and higher mortality than those in the low-risk group. Besides, the heatmap of the 5-lncRNA prognostic signature demonstrated that the expression levels of LINC02408 and ST8SIA6-AS1 were higher in the high-risk group, while the expression levels of LEF1-AS1, LINC01224, and MEF2C-AS1 were higher in the low-risk group. The Kaplan–Meier analysis and log-rank test were applied to compare the differences of OS between the low-and high-risk groups. As shown in Figure 2D, the patients’ OS in the high-risk group were significantly shorter than those in the low-risk group (p = 0.000049). Moreover, the time-dependent ROC curves were used to evaluate the sensitivity and specificity of the 5-lncRNA signature. The results revealed that the area under curve (AUC) at 3 and 5 years of the signature was 0.779 and 0.788, respectively (Figure 2G).
3.4 Validation of the 5-lncRNA prognostic signature
The test set and the entire set were adopted to validate the 5-lncRNA signature. In these two data sets, patients were divided into low- and high-risk groups using the same cutoff value defined in the training set. The distribution of risk score, patients’ survival status, and the heatmap of the 5-lncRNA prognostic signature in the test set and the entire set was consistent with the findings in the training set (Figure 2B, C). We also observed that the OS of patients in the low-risk group was significantly longer than that of patients in the high-risk group (all p < 0.005, Figure 2E, F). In addition, the time-dependent ROC curves of the two data sets showed that the lncRNA-based prognostic signature had good predictive accuracy (all AUC > 0.75, Figure 2H, I).
3.5 Prognostic assessment of the lncRNA-based signature and clinical characteristics
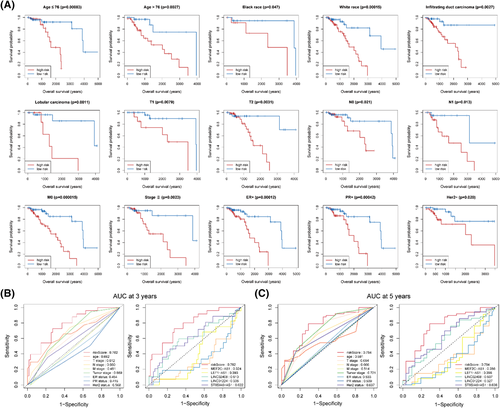
The univariate and multivariate Cox regression analyses were performed in the entire set to further assess the predictive value of the 5-lncRNA signature and clinical characteristics on the prognosis. Only the factors with a p-value < 0.1 in the univariate Cox regression analysis entered into the multivariate Cox regression analysis. The results showed that age, tumor stage, and 5-lncRNA signature were independent prognostic factors after multivariable adjustment of other clinical characteristics (Table 2). T stage and N stage were not included in the multivariate Cox regression analysis because these two factors are correlated with tumor stage, which can inevitably lead to multicollinearity and result in statistical bias.16 Furthermore, we performed the risk stratification analysis to evaluate whether our signature still could effectively predict the OS regardless of patients’ clinical characteristics. As shown in Figure 3A, patients in the high-risk group had significantly shorter OS than those in the low-risk group in different subgroups, including age ≤ 76 (median age, p = 0.00083), age > 76 (p = 0.0027), black race (p = 0.047), white race (p = 0.00015), infiltrating duct carcinoma (p = 0.0027), lobular carcinoma (p = 0.0011), T1 (p = 0.0079), T2 (p = 0.0031), N0 (p = 0.021), N1 (p = 0.013), M0 (p = 0.000015), tumor stage (p = 0.0023), ER + (p = 0.00012), PR + (p = 0.00042), and Her2- (p = 0.020). Furthermore, the time-dependent ROC analysis confirmed that the predictive accuracy of the 5-lncRNA signature was better than all the single genes and clinical characteristics (Figure 3B, C).
| Characteristics | Univariate Cox | Multivariate Cox | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Age | 1.134 (1.055–1.220) | 0.001 | 1.184 (1.058–1.325) | 0.003 |
| T stage | ||||
| T1 | 1.000 (Reference) | |||
| T2 | 1.819 (0.832–3.975) | 0.134 | ||
| T3 | 3.179 (1.239–8.157) | 0.016 | ||
| T4 | 1.999 (0.613–6.515) | 0.251 | ||
| N stage | ||||
| N0 | 1.000 (Reference) | |||
| N1 | 1.731 (0.821–3.648) | 0.149 | ||
| N2 | 3.043 (1.128–8.210) | 0.028 | ||
| N3 | 3.622 (1.155–11.357) | 0.027 | ||
| M stage | ||||
| M0 | 1.000 (Reference) | |||
| M1 | 2.446 (0.577–10.359) | 0.225 | ||
| Tumor stage | ||||
| Ⅰ | 1.000 (Reference) | 1.000 (reference) | ||
| Ⅱ | 2.104 (0.830–5.333) | 0.117 | 1.415 (0.383–5.219) | 0.603 |
| Ⅲ/Ⅳ | 3.594 (1.382–9.346) | 0.009 | 5.949 (1.425–24.831) | 0.014 |
| ER status | ||||
| Negative | 1.000 (Reference) | |||
| Positive | 0.688 (0.319–1.482) | 0.339 | ||
| PR status | ||||
| Negative | 1.000 (reference) | |||
| Positive | 0.579 (0.285–1.178) | 0.132 | ||
| HER2 status | ||||
| Negative | 1.000 (Reference) | 1.000 (reference) | ||
| Positive | 2.148 (0.888–5.194) | 0.090 | 1.231 (0.402–3.763) | 0.716 |
| Risk score | 1.670 (1.397–1.996) | <0.001 | 1.424 (1.100–1.844) | 0.007 |
Note
- The p value in bold indicates statistical significance (p < 0.05).
- Abbreviations: CI, confidence interval; HR, hazard ratio.
3.6 Nomogram development
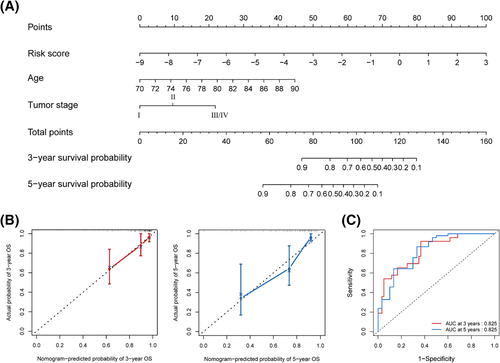
To quantitatively predict the overall survival probability of elderly patients with BC, we developed the prognostic nomogram based on the independent prognostic factors, including age, tumor stage, and the 5-lncRNA signature (Figure 4A). By using the generated nomogram, the total score for each patient was calculated to predict the probability of OS at 3 and 5 years. The calibration plots demonstrated high agreement between prediction and observation in 3- and 5-year overall survival probability (Figure 4B). The AUC for OS was 0.825 at 3 and 5 years (Figure 4C). Meanwhile, the C-index of the nomogram for OS prediction was 0.775, which was better than the C-index of single factor alone (age: 0.637; tumor stage: 0.667; 5-lncRNA signature: 0.744), indicating that the nomogram had excellent predictive accuracy in discriminating the patients with poor prognosis from those with good prognosis.
3.7 Enrichment analysis
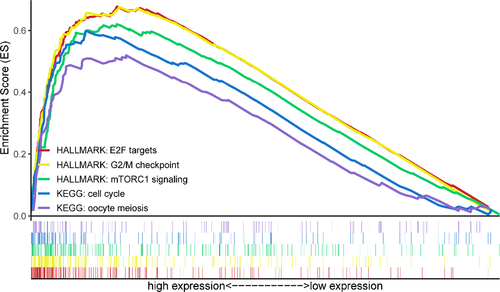
On the basis of the mRNA profile obtained from TCGA, GSEA was conducted to identify the potential molecular mechanisms of the 5-lncRNA signature in the prediction of elderly BC patients. The results revealed that three hallmark gene sets and two KEGG pathway gene sets were significantly enriched in the high-risk group, including E2F targets (NES = 1.91, FDR = 0.030), G2/M checkpoint (NES = 2.03, FDR = 0.011), mTORC1 signaling (NES = 2.18, FDR = 0.002), cell cycle (NES = 2.09, FDR = 0.021), and oocyte meiosis (NES = 2.12, FDR = 0.023). In contrast, none of the gene sets enriched considerably in the low-risk group (Figure 5).
4 DISCUSSION
With the aging of the population, we can expect that the proportion of elderly BC patients will probably increase significantly in the near future. However, due to the lack of sufficient clinical trial data, there are still no specific treatment guidelines for elderly BC, which often leads to undertreatment or overtreatment of patients and results in a poor prognosis.17 The available evidence suggests that elderly BC patients were treated less aggressively than their younger counterparts because of comorbidities and declining performance status.18 Therefore, it is of considerable significance to evaluate the prognostic risk of elderly patients with BC to guide the clinical treatment decision and improve patients’ overall survival.
LncRNA MALAT1 has been found to be markedly upregulated in BC tissues and may serve as an independent prognostic marker to predict the OS in BC patients diagnosed age below 60 or infiltrating ductal carcinoma.9 Li et al.10 constructed a 7-lncRNA model for the prognostic assessment of all types of BC patients with excellent predictive performance (AUC > 0.7). Another study showed that an 11-lncRNA based prognostic classifier was independent of traditional clinicopathological factors and was very reliable in predicting disease relapse of ER-positive BC patients who received tamoxifen therapy.11 However, it is not clear whether these prognostic biomarkers are applicable to the elderly, and lncRNAs have not been investigated as prognostic biomarkers in elderly BC patients to date.
In this study, the TCGA database, which contained large-scale genome sequencing data, comprehensive clinical information, and constantly updated follow-up records, was selected to construct the signature. First, we identified the aberrantly expressed lncRNAs associated with elderly BC by differential expression analysis. Then, through a combination of univariate Cox regression analysis, LASSO regression analysis, and multivariate Cox regression analysis, a 5-lncRNA signature to assess elderly BC patients’ prognosis was constructed in the training set. According to the median risk score, patients in the training set were successfully divided into a high-risk group with a poor prognosis and a low-risk group with a good prognosis. The 3- and 5-year AUC of time-dependent ROC curves were 0.779 and 0.788, respectively, indicating that the 5-lncRNA signature had a remarkable survival prediction ability. Additionally, the performance of our signature was validated in the test set and the entire set. Besides, univariate and multivariate Cox analyses revealed that the 5-lncRNA signature was an independent prognostic factor of OS for elderly BC patients. The risk stratification analysis showed that this signature could effectively predict patients’ prognosis with the same clinical characteristic. Moreover, to quantitatively and accurately evaluate the prognosis of elderly BC patients, we develop a prognostic nomogram on the basis of 5-lncRNA signature and patients’ clinical characteristics. The nomogram's calibration curves and C index indicated that the nomogram was an accurate predictor of predicting overall survival probability. It is worth noting that the 5-lncRNA signature was the dominant factor in this nomogram. Last but not least, through GSEA analysis, we revealed the underlying molecular mechanisms of our signature. The results showed that several pathways, such as E2F targets, G2/M checkpoints, mTORC1 signaling, cell cycle, and oocyte meiosis, were significantly enriched in the high-risk group, which may provide clues for clinical decision-making. MTORC1 signaling pathway participates in many age-related cancers and acts as a central regulator of angiogenesis, cell growth, and proliferation.19 Everolimus, an oral inhibitor of the mTOR pathway, exhibited promising anti-tumor activity in combination with trastuzumab in HER2 + metastatic BC patients and in combination with exemestane in HR + metastatic BC patients who had relapsed or progressed after receiving aromatase inhibitors.20 “E2F targets” and “G2/M checkpoints” are essential components involved in regulating cell cycle progression. Deregulation of the cell cycle is a characteristic of cancer that led to uncontrolled cell division. Cell cycle targeting agents such as cyclin-dependent kinase 4/6 (CDK4/6) inhibitors, and TTK protein kinase (TTK) and polo-like kinase 4 (PLK4) inhibitors, which are effective in the treatment of ER + BC and triple-negative BC, respectively, may benefit patients in the high-risk group.21 Oocyte meiosis is another cellular process associated with cell growth and death, but its role in BC has not been investigated.
Among the five lncRNAs in the signature, three of them, including MEF2C-AS1, LEF1-AS1, and LINC01224, are protective factors, while the other two lncRNAs (LINC02408 and ST8SIA6-AS1) acted as risk factors for BC. So far, some lncRNAs in our signature have been reported in the literature. Luo et al.22 illustrated that MEF2C-AS1 expression was significantly lower in gastric cancer tissues and patients’ plasma than in normal control. MEFC2-AS1 knockdown could promote aggressive tumor phenotypes and reduce the expression of FAT3, NTN1, and LYVE1, which were related to gastric cancer cell invasion and proliferation. Numerous studies have found that LEF1-AS1 was involved in the tumorigenesis and progression of different types of cancers. In colon cancer, LEF1-AS1 was upregulated and associated with poor prognosis, while knockdown of LEF1-AS1 suppressed cell migration, invasion, and metastasis through miR-30-5p/SOX9 Axis.23 In glioblastoma tissues, LEF1-AS1 was highly expressed and elevated cell malignant behaviors by sponging miR-543 to upregulate the expression of EN2.24 LEF1-AS1 was also overexpressed and functioned as an oncogene in hepatocellular carcinoma,25 ovarian cancer,26 retinoblastoma,27 osteosarcoma,28 esophageal squamous cell carcinoma,29 non-small-cell lung cancer,30 oral squamous cell carcinoma,31 and prostate cancer,32 while in myeloid malignancy, LEF1-AS1 was downregulated and served as a tumor suppressor lncRNA.33 LINC01224 in our signature has also been used to establish a risk model for predicting BC patients’ prognosis in a previous study.10 In a study of hepatocellular carcinoma, researchers demonstrated that LINC01224 facilitated tumor progression by upregulating CHEK1 expression through the competitive binding of miR-330-5p.34 Of the five lncRNAs in the prognostic signature, only ST8SIA6-AS1 has been researched in BC. Fang et al.35 found that ST8SIA6-AS1 expression in BC tissues was higher than that in adjacent normal tissues, and high expression of ST8SIA6-AS1 was correlated with ER-, PR-, advanced TNM stage, and poorer prognosis in BC patients. Moreover, the researchers speculated that ST8SIA6-AS1 might promote the proliferation, invasion, and migration of BC cells through the P38 MAPK signaling pathway. Luo et al.36 showed that ST8SIA6-AS1 was overexpressed in BC tissues, and high expression of ST8SIA6-AS1 was significantly associated with worse survival of BC patients, especially in the luminal subgroup. ST8SIA6-AS1 prevented tumor cells apoptosis by enhancing Aurora A-mediated PLK1 activation, loss of ST8SIA6-AS1 triggered mitotic catastrophe and massive cell death. Jeong et al.37 suggested that ST8SIA6-AS1 was upregulated in BC, mainly involved in increasing cell apoptosis and inhibiting cell migration, and was significantly associated with the regulation of the interferon signaling pathway during carcinogenesis. LINC02408, unfortunately, has not been researched up to now.
Although the 5-lncRNA signature is an ideal predictor of prognosis in elderly BC patients, some limitations should be taken into account. First, due to the limited number of elderly BC patients in the TCGA database and strict inclusion criteria, the sample size of this study was relatively small, which may lead to bias in our results. Therefore, large-scale, multicenter, prospective clinical trials are needed to validate this signature's performance before it can be applied to clinical practice. Second, the construction and validation of the 5-lncRNA signature were all carried out in the TCGA database, so external validation is required to evaluate our signature. Lastly, the molecular mechanism of the 5-lncRNA signature was inferred by bioinformatics analysis, and further in vitro and in vivo experiments are necessary to elucidate the inherent relationship between these five lncRNAs and the prognosis of elderly BC patients.
5 CONCLUSION
In summary, our results suggested that the 5-lncRNA signature is reliable in predicting the overall survival of elderly BC patients, which is an independent prognostic factor after multivariable adjustment of traditional clinical characteristics. The GSEA revealed the molecular mechanisms of the 5-lncRNA signature, providing several potential therapeutic strategies for the clinical treatment of patients in the high-risk group. Moreover, the nomogram constructed by independent prognostic factors such as 5-lncRNA signature, age, and tumor stage is a quantitative prediction tool that can more effectively assess the overall survival probability of elderly BC patients.
CONFLICT OF INTEREST
The author reports no conflicts of interest in this work.
AUTHOR CONTRIBUTIONS
Yi-Qun Che and Yang Luo conceived and designed the study. Yu-Xin Wu, Han-Bing Li, and Di Shen performed the literature search and data collection. Yue Zhang analyzed data and wrote the paper. All authors read and approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The RNA sequencing data and relevant clinical information were downloaded from TCGA database (http://cancergenome.nih.gov/).




