Circular RNA circβ-catenin aggravates the malignant phenotype of non-small-cell lung cancer via encoding a peptide
Weijun Zhao and Yandan Zhang contributed equally.
Funding information
This study was supported by Ningbo Public Welfare Science and Technology Plan Project (202002N3179).
Abstract
Background
More and more evidences demonstrate that circular RNAs (circNRAs) can encode protein. As a circRNA with translation capabilities, outcomes of circβ-catenin in non-small cell lung cancer (NSCLC) still need to be explored.
Method
The research methods of circβ-catenin in the article include qRT-PCR, wound healing assay, CCK-8, colony formation, and Transwell assay. Western blotting and immunofluorescence were provided to detect protein expression levels and peptide encoded by circβ-catenin, respectively.
Results
A prominently higher circβ-catenin expression was found in NSCLC tissues. Silencing of circβ-catenin was able to inhibit NSCLC cell migrating, invasive, and proliferative phenotypes. Overexpression of circβ-catenin could enhance the migrating, invasive, and proliferative phenotypes of NSCLC cells. Importantly, circβ-catenin was found to encode a peptide in NSCLC cells. Silencing or overexpression of circβ-catenin could reduce or increase β-catenin protein expression via suppressing the degradation of β-catenin.
Conclusion
Circβ-catenin could promote NSCLC cell malignant phenotypes via peptide-regulated β-catenin pathway. Our study provided a new understanding for the mechanisms of NSCLC.
1 INTRODUCTION
The publicly reported mortality rate associated with non-small-cell lung cancer (NSCLC) ranks fourth among all diseases.1 Although more and more patients benefit from surgery, radiotherapy and chemotherapy, the high rates of recurrence and metastasis still exist, and they are considered to be the main cause of death.2, 3 CircRNAs can be divided into noncoding circRNAs and coding circRNAs, which are considered to be endogenous small RNAs that are widely distributed, diverse in variety, and have multiple regulatory functions.4, 5 In 1976, 40 years ago, circRNAs were first discovered.6 In 1979 and 1986, it was confirmed that eukaryotic cells and humans also expressed circNRAs.7, 8 In 2012, Salzman et al. found that large number of genes can be transcribed to generate circRNAs.9
Abundant studies have shown that human diseases related to circRNAs include nervous, bone and joint, tumor, cardiovascular, endocrine, and respiratory diseases.10-17 CircRNAs also play a key role in cancer proliferation, metastasis, stem cell formation, and chemosensitivity.18, 19 As an RNA with complex functions, the endogenous RNA sponge of miRNA, regulation of gene expression, alternative splicing and molecular interaction, and the assembly of supporting protein complexes can all be accomplished through circRNAs.20, 21 Interestingly, more and more evidences demonstrate that circNRAs can encode protein. Legnini et al.22 found that an open reading frame (ORF) appearing in circZNF609 can encode protein in mouse myoblasts via being driven by internal ribosome entry site (IRES). In addition, circSHPRH23 and circFBXW724 and their encoded proteins are expressed in large amounts in nervous system, but are downregulated in gliomas. These circRNAs contain ORF of transcriptional functional proteins driven by IRES. Similarly, circMbl has been discovered to encode proteins by a cap-independent way.25
Circβ-catenin was reported to encode a novel peptide to promote the growth of liver cancer cell by β-catenin pathway.26 However, the role and mechanism of circβ-catenin in NSCLC remained unknown. In the present study, circβ-catenin was considered to promote the NSCLC cell malignant phenotypes via peptide-regulated β-catenin pathway. In the following, we will gradually explore the functions of circβ-catenin in NSCLC from shallow to deep.
2 METHODS AND MATERIALS
2.1 Patients and tissue samples
Cancer and paracancerous normal tissues from 20 patients with NSCLC were gained in the Ningbo First Hospital, Zhejiang, China. All patients signed an informed consent form. All protocols were agreed by the Ethics Committee of Ningbo First Hospital and in accordance with the 1964 Helsinki declaration.
2.2 Cell lines
BEAS-2B (Human normal lung epithelial cell line) and human NSCLC cell lines including H1299 and A549 were obtained from the American Type Culture Collection (ATCC). These cell lines were cultured in (Dulbecco's Modified Eagle Medium, Gibco) supplemented with 10% (Fetal Bovine Serum, Gibco) and 100 U/ml penicillin–streptomycin antibiotics (Gibco).
2.3 RNA isolation and real-time PCR
We extracted total RNA from NSCLC tissues, adjacent normal tissues and cells with TRIzol (Beyotime). The synthesis of cDNA was based on the instructions of PrimeScript RT Reagent Kit (Takara). ABI 7500 Fast Real-Time PCR System (Life Tech) and 2−ΔΔCt method were used to perform qRT-PCR and calculate gene expression respectively. The sequences of primers were listed: circβ-catenin (Forward, 5′-AGTGCTGAAGGTGCTATCTGT-3′; Reverse: 5′-AGGTAAGACTGTTGCTGCCA-3′), β-catenin (Forward, 5′- GGGTCCTCTGTGAACTTGCTC −3′; Reverse: 5′- TTCTTGTAATCTTGTGGCTTGTCC-3′) and GAPDH (Forward, 5′-CGCTCTCTGCTCCTCCTGTTC-30; Reverse, 50- ATCCGTTGACTCCGACCTTCAC-3′).
2.4 Western blotting and co-immunoprecipitation
IgG, HA, and GSK3β were added into lysis buffer gained from cells and incubated overnight at 4℃ with shaking. Then we mixed mix protein A/G PLUS-agarose (Beyotime) and cell lysates and incubated with shaking for 2 h. Lysis buffer was used to wash the agarose beads for 3 times. The final cell lysates were separated by Western blotting. Total proteins were obtained using RIPA buffer (Beyotime) and separated on 8%–12% SDS-PAGE. According to previous study,27 the PVDF membranes were probed by antibodies. The antibodies were listed as follow: β-catenin (66379–1-Ig, Proteintech, 1:1,000), Phospho-β-Catenin (#9561, Cell Signaling Technology, 1:1,000), GSK3β (67329–1-Ig, Proteintech, 1:1,000), HA tag (#5017, Cell Signaling Technology, 1:1,000), Vimentin (ab92547, Abcam, 1:1,000), E-cadherin (ab40772, Abcam, 1:1,000), and β-actin (AA128, Beyotime, 1:1,000). The expression levels were developed under enhanced chemiluminescence (ECL, Millipore).
2.5 Cell proliferation
A549 and H1299 cell proliferations were measured by CCK8 assays (Beyotime) according to previous study28; 100 microliters of 2,000 cells per well was added into 96-well plates. We added 10 μl CCK-8 solution to each well at each test time point and then kept for incubation for 1h. Finally, the absorbance was measured at 450 nm.
2.6 Colony formation
According to previous study,29 about 1,000 cells were added into 6-well plates and were grown for about 2 weeks. Cells fixed with 4% paraformaldehyde were stained by crystal violet for 10 min after being washed for 3 times with PBS.
2.7 Wound healing assay
The 100% confluent A549 and H1299 cells were cracked by the pipette tip. The distance of migration was assessed on Image J software (National Institutes of Health, USA) after 24 h later.
2.8 Transwell assay
A total of 100,000 cells in 100 μl serum-free DMEM were planted into the upper chamber coated with Matrigel in advance. After 24 h, cells fixed with 4% formaldehyde were stained with crystal violet for 15 min. The upper compartment cells were wiped by using a cotton swab. Finally, the relative numbers of invasion cells were calculated on Image J.
2.9 Plasmids, small interference RNAs and transfection
Circβ-catenin overexpressing vector (Lv-circβ-catenin), HA-Tag labeling circβ-catenin overexpressing vector (Lv-HA-circβ-catenin), HA-Tag labeling plasmid (Lv-HA-NC), and negative control plasmid (Lv-NC) were generated by Genomeditech, Shanghai, China. Circβ-catenin expression was instantly a knockdown by human circβ-catenin-specific small interference RNAs (siRNAs), which were generated from Genomeditech, Shanghai, China. Vectors were transfected with Lipofectamine 3,000 transfection reagent. After 48 h, cells were used to the follow-up experiments.
2.10 Immunofluorescence staining
Immunofluorescence staining was performed as done in previous study.30 H1299 cells were added onto glass coverslips in a 6-well plate. After Lv-HA-circβ-catenin transfection for 48 h, coverslips were fixed by 4% paraformaldehyde for 20 min. Cells permeabilized with 1% Triton X-100 were blocked with PBS containing 5% bovine serum albumin. Subsequently, HA primary antibody was added onto coverslips, incubated overnight at 4℃ and washed for 3 times the next day. Secondary antibody at room temperature was incubated for 1 h. Photos were taken by fluorescence microscopy (Olympus BX51).
2.11 Statistics
Mean ± SD was used to present the data and were calculated by GraphPad Prism 5.0 (GraphPad Software, Inc.). The significance of the difference between two groups were analyzed by the student's t-test. *p < 0.05, **p < 0.01, ***p < 0.001, versus the control was considered statistically significant. All experiments are repeated 3 times.
3 RESULTS
3.1 Circβ-catenin was involved in the migrating, invasive, and proliferative phenotypes of NSCLC cells
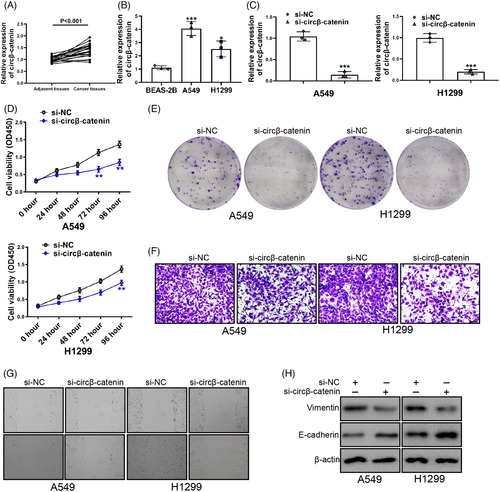
A prominently higher circβ-catenin expression was in cancer cell as previous study reported.26 First, we verified NSCLC cell circβ-catenin expression. An increased NSCLC cell circβ-catenin expression was discovered (Figure 1A). Next, we compared the circβ-catenin expression between BEAS-2B cells and NSCLC cells. Compared with BEAS-2B cells, circβ-catenin expression in A549 and H1299 cells increased by about 4 times and 2.5 times, respectively (Figure 1B). These results aroused our interest. In order to study the role of circβ-catenin in NSCLC, we synthesized small interference RNA to silence the expression of circβ-catenin (si-circβ-catenin) in NSCLC cells. Circβ-catenin expression was a knockdown by si-circβ-catenin (Figure 1C). Silencing of circβ-catenin reduced A549 and H1299 cells viability (Figure 1D). Silencing of circβ-catenin also inhibited NSCLC cell migrating, invasive, and proliferative phenotypes (Figure 1E, F, G). Importantly, silencing of circβ-catenin resulted in increased E-cadherin and decreased Vimentin expression in A549 and H1299 cells (Figure 1H). These results provided a point of view that circβ-catenin knockdown reduced the migrating, invasive, and proliferative phenotypes of A549 and H1299 cells via reducing epithelial cell-mesenchymal transition (EMT).
3.2 Upregulation of circβ-catenin promoted the malignant phenotypes of NSCLC cells
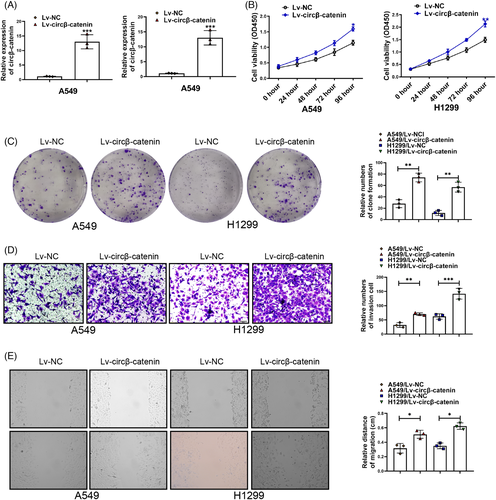
To further confirm the functions of circβ-catenin in NSCLC cells, Lv-circβ-catenin and Lv-NC were designed and synthesized. Upregulation of circRNAs can change the characteristics of tumor.31-34 The characteristics of tumor include many aspects, such as proliferation, migration, invasion, and drug resistance.28, 29, 35, 36 Therefore, we aimed to explore whether circβ-catenin upregulation could change the characteristics of NSCLC cells. Circβ-catenin expression was upregulated in A549 and H1299 cells transfected with Lv-circβ-catenin (Figure 2A). Circβ-catenin upregulation increased A549 and H1299 cells viability obviously (Figure 2B). Moreover, circβ-catenin upregulation also increased A549 and H1299 cell clone ability (Figure 2C). NSCLC cell migration and invasion were also promoted by circβ-catenin upregulation (Figure 2D, E). These results demonstrated that upregulation of circβ-catenin promoted the characteristics of NSCLC cells.
3.3 Circβ-catenin regulated the characteristics of NSCLC cells via Wnt/β-catenin pathway
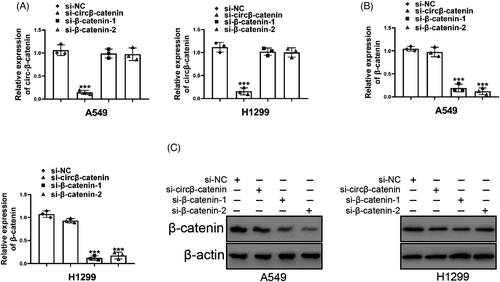
Since β-catenin was a cancer-promoting transcription factor of NSCLC and other cancers,37-42 we speculated that circβ-catenin in NSCLC cell might directly or indirectly affect β-catenin expression. To clarify whether there was an interaction between circβ-catenin and β-catenin, we designed and synthesized small interference RNA to knockdown β-catenin expression. As was shown in Figure 3A, B, si-circβ-catenin did not reduce β-catenin mRNA expression. β-catenin knockdown reduced the mRNA expression of β-catenin. Although si-circβ-catenin did not affect β-catenin mRNA expression, si-circβ-catenin reduced the protein level of β-catenin (Figure 3C). These results confirmed that circβ-catenin suppressed the characteristics of NSCLC cells via regulating Wnt/β-catenin pathway.
3.4 Circβ-catenin encoded a novel peptide in NSCLC cells
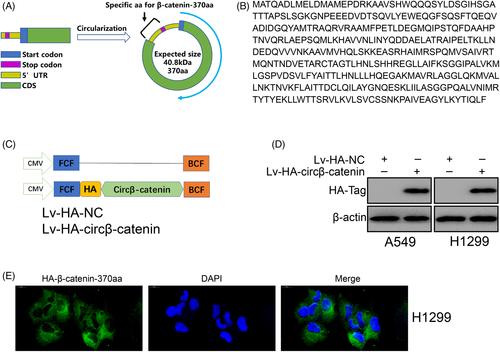
New evidences show that some circRNAs have protein-coding capabilities.23, 43, 44 CircSHPRH can encode a novel protein to inhibit glioma tumorigenesis.23 CircRNA LINC-PINT represses oncogenic transcriptional elongation in glioma via encoding a peptide.43 The analysis of circβ-catenin demonstrated that circβ-catenin had an ORF. According to previous study,26 circβ-catenin containing IRES encoded a peptide of 370 amino acids in size. In this article, this peptide was named “circβ-catenin-370aa”. The predicted molecular size of the peptide was 40.8 kDa (Figure 4A). An n-terminal sequence homologous to β-catenin was found on this peptide, and a C-terminal containing nine specific amino acids was found (Figure 4B). To verify circβ-catenin-370aa in NSCLC cells, we added a HA-Tag after the circβ-catenin-370aa coding sequence to label the circβ-catenin-370aa peptide (Figure 4C). Then we performed Western blotting and found that HA-Tag was shown on the membrane (Figure 4D). Immunofluorescence results show that circβ-catenin-370aa mainly existed in the cytoplasm (Figure 4E). These results confirmed that a novel peptide be translated from circβ-catenin in NSCLC cells.
3.5 Circβ-catenin-370aa inhibited β-catenin degradation via binding GSK3β
Since circβ-catenin knockdown did not decrease β-catenin mRNA expression but inhibited β-catenin protein level, we believed that circβ-catenin might reduce protein synthesis or degradation. β-catenin stability is believed to be related to its phosphorylation state. β-catenin phosphorylated by GSK3β can be ubiquitinated by β-TrCP and then degraded by the proteasome. Interestingly, β-catenin knockdown leads to a decrease in phosphorylated β-catenin (Figure 5A). β-catenin might be phosphorylated by GSK3β via interacting with its n-terminal. We speculated that β-catenin-370aa could prevent GSK3β from phosphorylating β-catenin by binding to GSK3β. According to the results of co-IP experiments, we found that there was a physical interaction between β-catenin-370aa and GSK3β (Figure 5B).
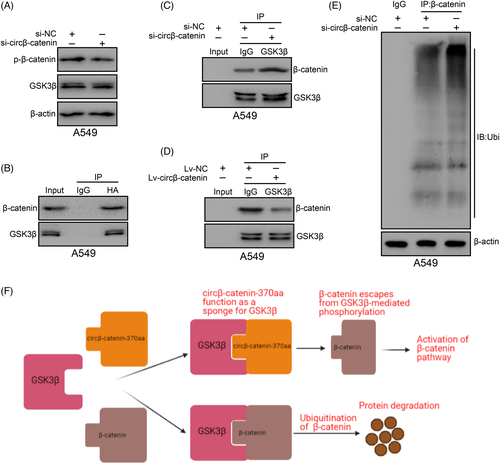
To further confirm the role of circβ-catenin in the reciprocity between β-catenin and GSK3β, circβ-catenin was a knockdown in A549 cells. Based on the results of co-IP experiments, circβ-catenin reduction significantly promoted the reciprocity between β-catenin and GSK3β in A549 cells (Figure 5C). However, circβ-catenin upregulation significantly reduced the reciprocity between β-catenin and GSK3β in A549 cells (Figure 5D). Considering that GSK3β-mediated β-catenin phosphorylation might trigger β-catenin ubiquitination, we knockdown circβ-catenin, used β-catenin antibody to pull down the β-catenin protein and then used ubiquitin antibody to detect its modification. According to the results of co-IP experiments, ubiquitinated β-catenin expression was increased obviously after knockdown of circβ-catenin (Figure 5E). These results showed that circβ-catenin increased β-catenin protein level by reducing its ubiquitination (Figure 5F).
4 DISCUSSION
In recent years, the human genome has been transcribed in large quantities, and abundant noncoding transcripts have been found in the human transcriptome.45 CircRNAs have received widespread attention in recent years.46, 47 The extensive expression patterns of circRNAs strongly illuminate their role in oncogenic regulation.33, 35, 46 CircRNAs are ubiquitous in the occurrence and development of NSCLC. For example, the expression of circSMARCA5 is negatively correlated with the tumor size of NSCLC, lymph node metastasis and TNM staging.48 CircRNA_103762 reduces the sensitivity of NSCLC to multi-drug chemotherapy by targeting DNA damage inducible transcript 3.49 Has_circ_0007385 expression was increased in NSCLC tissue and might be used as a new biomarker for disease monitoring and prognosis prediction of NSCLC patients.49 Recent study has revealed the role of a tumor promoter peptide encoded by the circβ-catenin in liver cancer.26 However, whether there is protein-encoded circRNA circβ-catenin involved in the occurrence of NSCLC tumors is still unknown.
In the current study, circβ-catenin expression was higher in NSCLC tumor tissue than paracancerous normal tissues. Overexpression or reduction of circβ-catenin remarkably enhanced or reduced the malignant characteristics of NSCLC cells. Subsequently, we found that circβ-catenin could be translated and encode a peptide in NSCLC cells. Interestingly, our findings demonstrated that circβ-catenin knockdown was able to reduce β-catenin protein expression in NSCLC cells. However, circβ-catenin knockdown could not decrease the mRNA level of β-catenin in NSCLC cells. Moreover, small interference to β-catenin was capable of reducing β-catenin mRNA and protein both. These results revealed that circβ-catenin knockdown affected the translation or degradation of β-catenin protein.
According to a previous study,26 circβ-catenin containing IRES sequence encoded a peptide of 370 amino acids in size. In this article, this peptide was named “circβ-catenin-370aa.” The predicted molecular size of the peptide was 40.8 kDa. An n-terminal sequence homologous to β-catenin was found on this peptide, and a C-terminal containing nine specific amino acids was found. Our results confirmed this point. We also found that circβ-catenin-370aa was mainly located in cytoplasm. Selective splicing of linear mRNAs offers the possibility of producing a lot of different protein subtypes from a limited number of protein-coding genes in the human genome.50 Most previous studies believed that circRNA cannot be transcribed and translated.51, 52 However, there is evidence that a small number of circRNAs can generate new protein subtypes via reverse splicing and circularization of selected exons in their linear mRNA transcription products. After the cycle, a new stop codon is generated and located upstream of the linear mRNA start codon AUG. After the translation of the circRNA, a new protein subtype containing additional amino acids will be produced. Previous studies have shown that 735 circRNAs have typical start codons as their linear counterparts.53
The n-terminal of this peptide was very similar to wild-type β-catenin. And our results showed that circβ-catenin knockdown was able to reduce β-catenin protein level but not mRNA level in NSCLC cells. Phosphorylated β-catenin is recognized and bound by β-Trcp and GSK3β.54 β-Trcp acts as an E3 ubiquitin ligase to target β-catenin for proteosome degradation,55 which prevents the regulation of downstream target genes. Therefore, we speculated that circβ-catenin-370aa, a novel peptide, circβ-catenin replaces β-catenin by deceiving and binding to GSK3β and prevents the degradation of β-catenin. co-IP results confirmed that circβ-catenin knockdown or overexpression was able to enhance or reduce the combination between GSK3β and β-catenin. In addition, it is known that β-catenin can increase the malignant phenotype of NSCLC.41, 42, 56
In summary, these results indicated that circβ-catenin encoded β-catenin-370aa to combine with GSK3β, leading to escape the degradation of β-catenin induced by GSK3β.
CONFLICT OF INTEREST
All authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Yonggang Zhu carried out the experiments and collected data. Weijun Zhao conceived and designed the study. Yandan Zhang drafted the manuscript. All authors read and approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All data are included in this article.




