Ubiquilin proteins regulate EGFR levels and activity in lung adenocarcinoma cells
Zimple Kurlawala and Kumar Saurabh contributed equally to this study.
Abstract
Ubiquilin (UBQLN) proteins are involved in diverse cellular processes like endoplasmic reticulum-associated degradation, autophagy, apoptosis, and epithelial-to-mesenchymal transition. UBQLNs interact with a variety of substrates, including cell surface receptors, transcription factor regulators, proteasomal machinery proteins, and transmembrane proteins. In addition, previous work from our lab shows that UBQLN1 interacts with insulin-like growth factor receptor family members (IGF1R, IGF2R, and INSR) and this interaction regulates the activity and proteostasis of IGFR family members. We wondered whether UBQLN proteins could also bind and regulate additional receptor tyrosine kinases. Thus, we investigated a link between UBQLN and the oncogene epidermal growth factor receptor (EGFR) in lung adenocarcinoma cells. Loss of UBQLN1 occurs at high frequency in human lung cancer patient samples and we have shown that the loss of UBQLN1 is capable of altering processes involved in cell proliferation, migration, invasion, and epithelial-to-mesenchymal transition in lung adenocarcinoma cell lines. Here, we present data that loss of UBQLN1 resulted in increased turnover of total EGFR while increasing the relative amount of phosphorylated EGFR in lung adenocarcinoma cells, especially in the presence of its ligand EGF. Furthermore, the loss of UBQLN1 led to a more invasive cell phenotype as manifested by increased proliferation, migration, and speed of movement of these lung adenocarcinoma cells. Taken together, UBQLN1 regulates the expression and stability of EGFR in lung cancer cells.
Abbreviations
-
- AD
-
- Alzheimer's disease
-
- BCL2
-
- B-cell lymphoma 2
-
- EGF
-
- epidermal growth factor (ligand)
-
- EGFR
-
- epidermal growth factor receptor
-
- EMT
-
- epithelial-to-mesenchymal transition
-
- IGFR
-
- insulin-like growth factor receptor
-
- NSCLC
-
- non–small cell lung cancer
-
- RTK
-
- receptor tyrosine kinase
-
- UBA
-
- ubiquitin-associated domain
-
- UBL
-
- ubiquitin-like domain
-
- UBQLN
-
- ubiquilin
1 INTRODUCTION
Cancer and Alzheimer's disease (AD) are seemingly caused by contrasting cellular processes: aberrant cell survival for cancer and aberrant cell death for AD.1 The family of adapter proteins, ubiquilins (UBQLNs), are lost in multiple types of cancers as well as in AD.2-4 This family consists of five members; UBQLN1-UBQLN4 and UBQLNL, which contains an n-terminus ubiquitin-like (UBL) domain and a c-terminus ubiquitin-associated (UBA) domain.5-7 UBQLN1 is involved in a variety of cellular processes like endoplasmic reticulum-associated degradation,8, 9 autophagy,10, 11 apoptosis,12 and epithelial-to-mesenchymal transition (EMT).9, 13 UBQLN1 also interacts with diverse substrates—proteins involved in the proteasomal machinery (PSMD4 and BAG6),6 cell surface receptors GABA-A,14 GPCR's,15 PSEN1/2,16, 17 and IGF1R,6, 18 transcription factor regulators IκBα,19 and other transmembrane proteins ESYT2,6 CD47,20 and BCLb.2 UBQLN1 is a versatile multipurpose adaptor that interacts with a wide range of substrates; thus, it can regulate multiple important cellular processes.
UBQLN2, is another UBQLNs family member that is constitutively expressed in most cell types and shares more than 75% homology with UBQLN1, indicating that it likely shares similar biological functions.7 Like UBQLN1, UBQLN2 also has a UBL domain that interacts with the proteasome and a UBA domain which recognizes ubiquitin on target proteins.21, 22 In addition, UBQLN2 has a 12-PXX repeat region which makes it unique among the UBQLN family proteins.22
Receptor tyrosine kinases (RTKs) are cell surface receptors found to be responsible for mediating signaling pathways crucial to cell proliferation, cell migration, and invasion of many types of cancer.23 Our lab was first to identify the interaction of UBQLN1 with an RTK family member, namely insulin-like growth factor receptors (IGFRs).18 Loss of UBQLN1 leads to a significant decrease in the amount of total IGF1R, an increase in phosphorylated IGF1R, and dramatic increases in their migratory potential when stimulated with IGF in lung adenocarcinoma. Epidermal growth factor receptor (EGFR), a transmembrane protein that is also a member of the RTK family, is mutated or overexpressed in multiple cancers and AD.24-26 EGFR is one of the most commonly studied oncogenes to date. It is often upregulated in multiple cancers and downregulated in AD.1 This unique inverse relationship between cancer and AD has been classified for a wide array of proteins, including p53, IGF1R, and B-cell lymphoma 2 (BCL2).1 EGFR is a well-established therapeutic target of kinase inhibitors (e.g. gefitinib, erlotinib, and lapatinib) and monoclonal antibodies (e.g. cetuximab, panitumumab, and trastuzumab). Many of these therapeutics are now being used as first-line treatment for both lung cancer and Alzheimer's patients.24-26 In this study, we present data establishing the interaction of UBQLN1 and UBQLN2 with EGFR. In lung adenocarcinoma cell lines, downregulation of UBQLN1, followed by epidermal growth factor (EGF) stimulation, leads to degradation of total EGFR protein, and an increase in migration and invasion potential of these cells.
2 MATERIALS AND METHODS
2.1 Cell culture, transfection, and EGF stimulation
Human embryonic kidney 293T (HEK293T) cells were procured from American Type Culture Collection (ATCC, Rockville, MD) and cultured in Dulbecco's modified Eagle's medium (#SH30243; Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS; #SH30070; Hyclone) and 1% antibiotic/antimycotic (#SV30010, Hyclone) at 37°C with 5% CO2. A549 (lung adenocarcinoma line) were procured from ATCC and cultured in RPMI (#SH30027; Hyclone) supplemented with 10% FBS and 1% antibiotic/antimycotic. Small interfering RNA (siRNA) transfections were performed as described previously.18 Briefly, 48 hours after transfection, cells were serum starved (SS) for 3 hours, incubated with protein synthesis inhibitor cycloheximide (CH, 10 µM) for 1 hour before supplementing serum-free (SF) media with 25 or 50 ng/mL EGF (#PHG0314; Thermo Fisher Scientific, Waltham, MA), as indicated in the experiments. Three hours after ligand stimulation, cells were harvested and probed for EGFR (total and phosphorylated) protein by Western blot analysis. For dose and time-dependent studies, cells were stimulated and/or incubated for doses and time points indicated in respective figures. Densitometry analyses of Western blot band images were performed using NIH ImageJ software, following the steps described here http://www.navbo.info/DensitometricAnalysys-NIHimage.pdf.
2.2 Plasmid construction
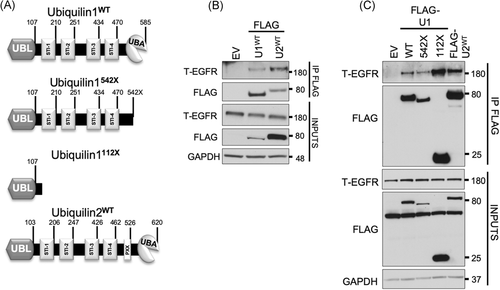
As described previously, constructs with deleted domains of UBQLN1 (Figure 1A) were developed using Q5 Site-Directed Mutagenesis Kit as per manufacturer's protocol (#E0554; New England Biolabs, Ipswich, MA) and confirmed by sequencing.6, 27
2.3 Immunoprecipitation, protein estimation, and Western blot
Immunoprecipitation (IP) was performed as described previously.6 Briefly, harvested cells for each procedure (IP and/or transfection) were lysed with 1% CHAPS lysis buffer and total protein was estimated using the bicinchoninic acid quantification method.6 Western blot analyses were performed in Bolt Bis-Tris gels (#BG4120BOX; Life Technologies, Grand Island, NY) as per manufacturer's protocol using antibodies GAPDH #sc47724 (Santa Cruz, Dallas, TX); Actin #A5316 (Sigma-Aldrich, St. Louis, MO); UBQLN polyclonal was made by inoculating rabbits with a peptide specific to UBQLN1 (Yenzym Antibodies LLC, South San Francisco, CA); and UBQLN1 #14526, FLAG #14793, EGFR #4267, and pEGFR Tyr1068 #3777 (Cell Signaling, Danvers, MA).
2.4 Cell viability and migration assay
Cell viability and migration assays were performed as described earlier.7 Briefly, A549 cells were cultured in 60-mm culture plates. After 12 hours of transfection with siRNA, cells were trypsinized, counted, and 2000 cells were reseeded per well in 96-well plates in complete media. Twelve hours after reseeding in complete media, cells were SS for 3 hours followed by stimulation with EGF, and were cultured in media containing 2% FBS. Cell viability was analyzed for four successive days using AlamarBlue (#DAL1100; Thermo Fisher Scientific). At the same time following transfection, 5000 cells were seeded in Transwell cell culture inserts (#CLS3464; Corning Inc, Corning, NY) in triplicate for each condition as described previously.9 Briefly, cells were allowed to grow on Transwell cell culture insert in SF media, SF media supplemented with EGF (50 ng/mL), and SF media supplemented with both EGF and erlotinib (1 µM). After 24 hours, membranes were washed once with PBS, fixed with ethanol, stained with Giemsa stain (#R03055; Sigma-Aldrich), and cells were counted on the microscope.
2.5 Live cell imaging
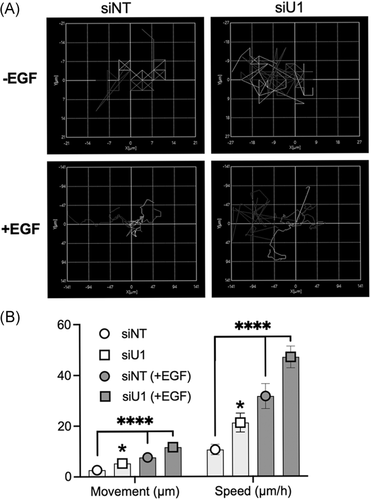
A549 cells were transfected with siRNA against UBQLN1 and non-targeting (NT) control. Twenty-four hours after transfection, cells were trypsinized, counted, and 10 000 cells were reseeded per well in 12-well plates, coated with thin layer commercial extracellular matrix (ECM; #E6909; Sigma-Aldrich) at a concentration of 1 µg/cm2 for 12 hours (overnight) in complete media. The next day, all wells were SS for 3 hours. Cells were then maintained in media with 2% FBS or 2% media supplemented with EGF (25 ng/mL). Cells were imaged for 48 hours on a time interval of 15 minutes on Keyence BZ-X810. All pictures were stitched together to produce a video with a speed of 14 fps. For dynamic tracking, five single cells were analyzed on Keyence BZ-X810 software to generate a Chemotaxis plot and to calculate the movement and speed of cells.
2.6 RNA interference sequences
All RNA interference (siRNAs) used for the study were ordered from Thermo Fisher Scientific Biosciences Inc (Lafayette, CO) and transfections were done using Dharmafect1 as per the supplier's instructions. (a) NT control, UAAGGCUAUGAAGAGAUACAA and (b) UBQLN1, siU11: GAAGAAAUCUCUAAACGUUUUUU and siU12: GUACUACUGCGCCAAAUUUUU.
2.7 Statistical analysis
All statistics were performed using GraphPad Prism 8 software. Unless otherwise specified, significance was determined by one-way or two-way analysis of variance, using a cut off of P < .05.
3 RESULTS
3.1 UBQLN1 and UBQLN2 interact with EGFR
Our previous data described that UBQLN1 interacts with IGF1R. In this study, we aimed to explore the possibility that UBQLN proteins might interact with and regulate additional RTK family member, EGFR. We first performed a co-IP of UBQLNs to identify interacting proteins (Figure 1). We transiently transfected HEK293T cells with UBQLN1-FLAG or UBQLN2-FLAG constructs, then pulled down UBQLN-interacting proteins using anti-FLAG-conjugated agarose beads. Western blot analysis of these FLAG-IP samples revealed that UBQLN1 and UBQLN2 interacted with total EGFR (Figure 1B). Like UBQLN1, UBQLN2 has an n-terminus UBL, a c-terminus UBA, and four STI (STI1-STI4) domains, plus UBQLN2 has an additional unique PXX (12 tandem repeats) domain (Figure 1A). To determine the interacting domain of UBQLN1, we created two constructs as described in Figure 1A and performed a similar co-IP experiment. For one construct, we deleted the UBA domain of UBQLN1 (labeled 542X), and for the second construct, we deleted the UBL domain (labeled 112X). Our results indicated that the UBL domain is the primary site of interaction with EGFR. The deletion of the UBA domain did not cause a loss of interaction between UBQLN1 and EGFR (Figure 1C).
3.2 UBQLN1 regulates expression and activity of EGFR
Next, we investigated whether the loss of UBQLN1 regulated EGFR expression and activity in lung adenocarcinoma cells (Figure 2). A549 cells were transiently transfected with NT siRNA and two UBQLN1-specific siRNAs (Figure 2A). Cells were then SS to synchronize the cell cycle and remove all confounding growth factors present in serum. Then, cells were treated with CH to prevent the synthesis of new EGFR protein. Upon loss of UBQLN1, there was a decrease in total EGFR expression compared to the NT control. This decrease in total EGFR expression was significantly enhanced when the receptor was stimulated with EGF ligand. Next, we tested the regulation of total EGFR when stimulated with different doses of EGF ligand (Figure 2B). There was significantly increased degradation of total EGFR in cells lacking UBQLN1 compared to controls at 10 ng/mL, which was enhanced at 100 ng/mL. Interestingly, the expression of phosphorylated-EGFR (p-EGFR) was not as sensitive to the loss of UBQLN1. Therefore, in cells lacking UBQLN1, there was an EGF dose-dependent degradation of EGFR.
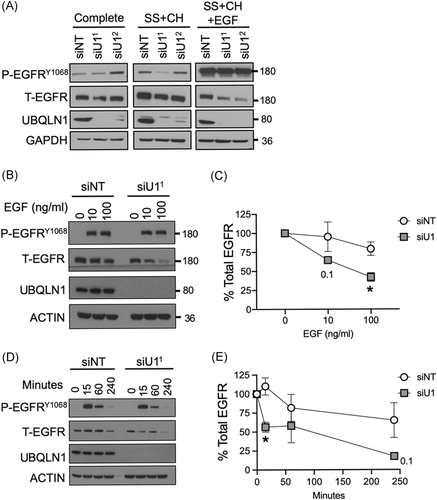
Next, we performed experiments to determine the temporal regulation of EGFR by UBQLN1 (Figure 2D,E). Lung adenocarcinoma cells were transfected with NT or UBQLN1-specific siRNAs. Two days posttransfection, cells were SS for 3 hours, then stimulated with EGF (50 ng/mL) for indicated time points. Upon stimulation with EGF, we observed degradation of total EGFR as time progressed. However, in cells lacking UBQLN1, there was significantly increased degradation compared to control as time progressed.
3.3 UBQLN1-deficient cells exhibit increased cell viability and migration potential
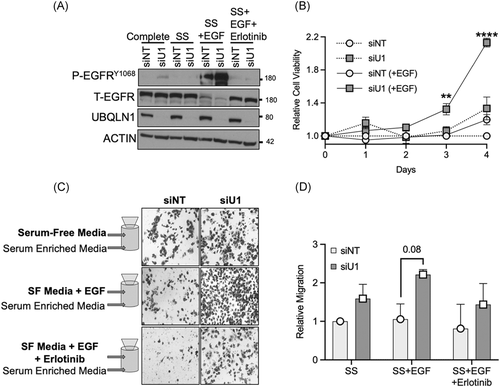
EGFR proteins play a role in maintaining cell viability and stimulating the migratory potential of lung adenocarcinoma cells. We investigated the influence of loss of UBQLN1 on these EGFR-mediated processes (Figure 3). A549 cells were transiently transfected with siRNA for UBQLN1 and NT control and cultured in different conditions as indicated (complete media, SS for 3 hours, SS + EGF [50 ng/mL], and SS + EGF + erlotinib, a p-EGFR inhibitor [1 µM]). Cells were harvested after 3 hours and analyzed by Western blot. When stimulated with EGF, UBQLN1-deficient cells showed almost complete loss of total EGFR and increased expression of p-EGFR, which was blocked by erlotinib (Figure 3A). Next, A549 cells were transfected with NT or UBQLN1-specific siRNAs, and cell viability was measured for 4 consecutive days using alamarBlue (Figure 3B). Consistent with our previous findings, loss of UBQLN1 resulted in increased cell growth in lung adenocarcinoma cell lines. Interestingly, there was an increase in the relative number of cells in UBQLN1-deficient cells stimulated with EGF, compared to controls. Next, we determined the effects of loss of UBQLN1 on cell migration (Figure 3C,D). Using a Transwell migration plate, we seeded A549 cells that had been transfected with either NT or UBQLN1-specific siRNAs and cultured the cells under three conditions—SF media; SF media supplemented with EGF; and SF media supplemented with EGF and erlotinib, a p-EGFR inhibitor. Consistent with our previous data using this migration model, UBQLN1-deficient cells exhibited an approximately threefold increase in migration when stimulated with EGF. Erlotinib decreased the migration of cells in both control and UBQLN1-deficient cells.
3.4 Loss of UBQLN1 results in increased cell movement and speed
We examined individual cell movement and speed to explore the increased migratory potential in cells lacking UBQLN1 (Figure 4). We utilized live-cell imaging equipped with image analysis software (Keyence). A549 lung adenocarcinoma cells were transfected with NT or UBQLN1-specific siRNAs for 24 hours after which they were SS for another 24 hours. At this point, 10 000 cells per well (12-well plates) were seeded on a thin layer of commercial ECM in the presence or absence of EGF (25 ng/mL) to yield the following conditions: siNT (+/−) EGF and siUBQLN1 (+/−) EGF. Live-cell images were captured for 48 hours using a Keyence microscope. All captured images were stitched together to make a representative video (Figure S1). As evident from the videos, loss of UBQLN1 resulted in increased movement of A549 cells which was further enhanced by EGF stimulation. These data are represented as chemotaxis plots (Figure 4A). In addition, we quantified the distance traveled and the rate of travel (speed) for each cell (Figure 4B). These data show that cells lacking UBQLN1 travel further and move faster as compared to the NT controls. These differences were enhanced in the presence of EGF.
4 DISCUSSION
UBQLN1 was reported to be lost in approximately 50% of non–small cell lung adenocarcinomas.9 Our group is interested in understanding how the loss of function of UBQLN proteins contributes to the metastatic progression of human lung adenocarcinoma.9, 13, 18 Previously, we have shown that interaction between UBQLN1 with IGF1R results in stabilization of this receptor. When UBQLN1 is lost, it leads to increased phosphorylation of the autophosphorylation site on the IGF receptor, while total IGF1R levels decreased. Similarly, in this study, we report that the loss of UBQLN1 does not alter p-EGFR expression while causing a robust decrease in total EGFR expression. In addition, we have reported dose and time dependent decreases in total IGF1R when stimulated with IGF ligand.18 Likewise, in this study, lung adenocarcinoma cells lacking UBQLN1 also exhibit an enhanced degradation of EGFR when stimulated with EGF that is both dose and time dependent. These data suggest that loss of UBQLN1 accelerates both EGFR and IGF1R turnover in cancer cells. UBQLN1 might be crucial to maintain the stability of these RTKs,28 thus influencing progrowth and prosurvival signaling pathways in lung adenocarcinoma cells.
We have previously demonstrated that the downregulation of UBQLN1 leads to significantly increased expression of mesenchymal markers like Vimentin, Snail, and ZEB1 indicating that UBQLN1 may play a role in the suppression of metastasis in lung cancer.9 In addition, knockdown of UBQLN1 by siRNA or mir155-mediated downregulation of UBQLN1 in lung cancer cells promoted an EMT-like phenotype.13 Data presented here further support the role of UBQLN1 in proliferation and migration of cancer cells and this was exacerbated in the presence of EGF stimulation. These results, when considered in conjunction with our previous work with IGF1R, suggest that cells lacking UBQLN1 then stimulated with IGF or EGF, enhanced the metastatic potential of cancer cells.18 Our results point to the destabilization of these RTKs when UBQLN1 is lost which further leads to an invasive phenotype in lung adenocarcinoma cells. In a related study, UBQLN4, another member of the UBQLN family, was shown to interact with RNF11, an E3 ubiquitin ligase of p21. Overexpression of UBQLN4 induced cellular senescence and cell cycle arrest in gastric cancer cells.29 These findings further support the significance of UBQLN proteins in cancer progression and tumorigenesis. As reported here, cells that lack UBQLN1 show increased cell movement and speed captured with live-cell imaging over multiple days. The overall movement and speed of these lung adenocarcinoma cells were further accelerated in the presence of EGF stimulation. RTK family of kinases plays critical roles in progression of human lung cancer. In non–small cell lung cancer (NSCLC), IGFR and EGFR are overexpressed and UBQLNs are underexpressed and synergistically contribute to tumor development and progression.30, 31 Collectively, these data indicate a critical role for UBQLNs in the normal proteolytic degradation of RTKs, and loss of UBQLN function in cancer cells leads to aberrant RTK-mediated signaling, further enhancing the metastatic potential of these cancers. Downstream pathways activated by these cell surface receptors crosstalk and upon mutual activation lead to acquired resistance against EGFR-targeted drugs. We propose that targeting both EGF and IGF receptors at once might enhance antitumor efficacy and would be a promising approach for NSCLC therapies.30-32 Being able to target both of these pathways simultaneously via their interaction with UBQLNs is an exciting avenue to explore.
Our lab has also been interested in the inverse relationship between cancer incidences and many neurodegenerative disorders. Data suggest that loss of UBQLN function may be involved in both diseases. In fact, after UBQLN1 was found to interact with the presenilin complex via a yeast-two hybrid screen, research efforts were quickly underway to establish the role that UBQLN family members might play in neurodegenerative diseases.3, 16, 22, 27, 33, 34 Single-nucleotide polymorphisms in the UBQLN1 gene are associated with late-onset of AD.33 In addition, mutations found in UBQLN2 drive early-onset amyotrophic lateral sclerosis/frontotemporal dementia.22, 34 Perhaps, UBQLN family members inversely regulate outcomes in cancer and neurodegenerative diseases such that loss of function of UBQLN proteins in epithelial cells leads to a cancerous phenotype, while in postmitotic neurons leads to a degenerative phenotype. Further experiments would be needed to establish the exact role.
Interestingly, some members of the BCL2 family, IGF1R, and EGFR are overexpressed in cancer but underexpressed in brains of patients with AD.1 Interestingly, each of these proteins, either in this study or in our previous work, has been shown to be stabilized by UBQLN1.2, 28 In AD, β-amyloid aggregations accumulate in the brain. Studies have shown that inhibiting IGF1R and EGFR leads to a significant reduction in β-amyloid aggregations.35, 36 Polyubiquitination of EGFR regulates its cellular location and stability, and this ubiquitination can be decreased by mutating the lysine residues without having an effect on the tyrosine kinase activity of EGFR.37 Thus, inhibition of RTKs could not only benefit certain cancers, but it could also be beneficial in several neurodegenerative diseases.26, 35, 36 These studies further support our belief that UBQLNs regulate many processes involved in multiple diseases.
In conclusion, the UBA-UBL domain-containing family of UBQLNs facilitates normal proteasomal degradation, and substrate selection by UBQLNs is critical in cancer and neurodegenerative diseases. Our IP data provide evidence for the interaction of both UBQLN1 and UBQLN2 (shares more than 75% homology with UBQLN1) with EGFR. Taken together with our previous findings, both IGFRs and EGFR were detected as interacting partners of UBQLN1. The lung adenocarcinoma cells lacking UBQLN1 exhibit enhanced degradation of both EGFR and IGF1R, especially in the presence of their respective ligands. Thus, UBQLN1 is important in maintaining the stability of RTKs, like EGFR and IGF1R. Loss of UBQLN1 alters the normal degradative fate of these receptors leading to downstream alterations in signaling that result in a cellular phenotype with enhanced proliferation, migration, and movement of the cells. The RTK family of proteins is highly regulated in both cancer and neurodegenerative diseases and this sets the stage for the development of directed therapies. Our work with UBQLNs indicates that they are uniquely poised as therapeutic targets given their role in regulating the stability of multiple members of the RTK family, and this regulation might be inversely impacted in cancer vs neurodegenerative diseases.
ACKNOWLEDGMENTS
The authors are grateful to the Beverly and Siskind lab members, especially Lavona Casson and Mark Doll, for continuous support and sharing of resources and insight. This study was supported by NIH R01CA193220, Kentucky Lung Cancer Research Program, and funds from James Graham Brown Cancer Center, University of Louisville to Levi J. Beverly. Rain Dunaway was supported by NIH National Cancer Institute Cancer Education Program Grant R25-CA134283.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
KS, ZK, RD, and PPS did the experiments. KS, ZK, LJS, and LJB conceived studies, did the analysis, and wrote the manuscript. All authors have read and approved the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.