Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation
ABSTRACT
Osteogenic lineage commitment and progression is controlled by multiple signaling pathways (e.g., WNT, BMP, FGF) that converge on bone-related transcription factors. Access of osteogenic transcription factors to chromatin is controlled by epigenetic regulators that generate post-translational modifications of histones (“histone code”), as well as read, edit and/or erase these modifications. Our understanding of the biological role of epigenetic regulators in osteoblast differentiation remains limited. Therefore, we performed next-generation RNA sequencing (RNA-seq) and established which chromatin-related proteins are robustly expressed in mouse bone tissues (e.g., fracture callus, calvarial bone). These studies also revealed that cells with increased osteogenic potential have higher levels of the H4K20 methyl transferase Suv420h2 compared to other methyl transferases (e.g., Suv39h1, Suv39h2, Suv420h1, Ezh1, Ezh2). We find that all six epigenetic regulators are transiently expressed at different stages of osteoblast differentiation in culture, with maximal mRNAs levels of Suv39h1 and Suv39h2 (at day 3) preceding maximal expression of Suv420h1 and Suv420h2 (at day 7) and developmental stages that reflect, respectively, early and later collagen matrix deposition. Loss of function analysis of Suv420h2 by siRNA depletion shows loss of H4K20 methylation and decreased expression of bone biomarkers (e.g., alkaline phosphatase/Alpl) and osteogenic transcription factors (e.g., Sp7/Osterix). Furthermore, Suv420h2 is required for matrix mineralization during osteoblast differentiation. We conclude that Suv420h2 controls the H4K20 methylome of osteoblasts and is critical for normal progression of osteoblastogenesis. J. Cell. Biochem. 118: 1262–1272, 2017. © 2016 Wiley Periodicals, Inc.
The epigenetic landscape of chromatin is shaped by regulatory enzymes and scaffolding proteins that are responsible for writing, reading, interpreting, editing, and/or erasing post-translational modifications on histone proteins [Ruthenburg et al., 2007]. These enzymes may function as pharmacological targets and thus present an enticing opportunity for manipulation of the biological properties of cells during cell growth and differentiation. Recently, we have shown that natural compounds can alter chromatin via active DNA demethylation [Thaler et al., 2012, 2016] and that drug-induced inhibition of the H3K27 methyltransferase Ezh2 significantly supports osteoblastic differentiation and bone formation in vitro and in vivo [Dudakovic et al., 2015, 2016]. These studies provide proof-of-concept that the activity of epigenetic regulators (EpiRegs) can be leveraged to promote osteogenic lineage progression [Fu et al., 2013; Hemming et al., 2014; Sabbattini et al., 2014], and provide the rationale for investigating the roles of other EpiRegs during osteoblast differentiation.
Transcriptional modulation of select differentiation-related target genes is in principle possible by inhibiting the deposition of active or repressive histone marks. Transcriptionally repressive methyl marks are found on lysine residues at position 9 and 27 on histone H3 and on position 20 on histone H4. The genome-wide presence of these marks in mesenchymal stromal cells and phenotype-committed osteoblasts is controlled by transcription factors that selectively recruit EpiRegs. For example, the essential osteoblast-related transcription factor Runx2, nuclear receptors (e.g., vitamin D receptor, Vdr), as well as other transcription factors were shown to modulate several epigenetic mechanisms during osteoblastogenesis [Meyer et al., 2014; Seth-Vollenweider et al., 2014; St John et al., 2014; Li et al., 2016]. Furthermore, our group and others recently elucidated how formation of the repressive mark histone 3, lysine 27, trimethylation (H3K27me3) by Ezh2 affects osteoblastic differentiation [Gordon et al., 2015; Dudakovic et al., 2015, 2016; Aguilar et al., 2016].
Many studies have focused on the three principal methylated lysines of histone H3 (i.e., H3K4, H3K9, and H3K27). Yet, less is known about the role of methylation at histone H4 lysine 20 (i.e., H4K20me1, H4K20me2, and H4K20me3) even though it plays a significant role in cell proliferation [Jorgensen et al., 2013], and thus is likely to control the transition from cell growth to differentiation. Mono-methylation on Histone H4 lysine 20 methylation (H4K20me1) is associated with transcriptional activation and regulates chromatin condensation and mitotic progression [Beck et al., 2012]. Conversely, tri-methylation on Histone H4 lysine 20 methylation (H4K20me3) is a hallmark of silenced heterochromatic regions, as well as related to repression of transcription and the activity of transposons [Schotta et al., 2004].
Enzymes involved in the methylation of H4K20 are represented by two distinct classes of histone methyltransferases related to the fruit fly “suppressor of variegation” (Suv) proteins, including Suv39 homologues 1 and 2, as well as Suv420 homologues 1 and 2 (respectively, Suv39h1, Suv39h2, Suv420h1, and Suv420h2) [Bradley et al., 2015; Gordon et al., 2015]. SUV proteins utilize the conserved SET domain to methylate lysines on side chains of histone tails. For example, SUV420H2 targets lysine 20 on the histone 4 N-terminal tail (H4K20) [Wu et al., 2013], while both SUV420H2 and SUV420H1 recognize the mono-methylated version of H4K20 (H4K20me1) to catalyze the addition of a second methyl group resulting in di-methylated H4K20 (H4K20me2) [Southall et al., 2014]. The substrate specificity of SUV420H2 has been shown to be highly specific for H4K20me1 [Yang et al., 2008]. From genetic and developmental perspectives, global loss of SUV420H1/2 results in perinatal lethality, indicating that these enzymes are principal regulators of normal embryonic patterning, as well as perhaps in self-renewal or lineage-commitment of stem cell populations [Schotta et al., 2008]. Indeed, SUV420H2 activity has also been shown to participate in myogenic lineage commitment [Tsang et al., 2010; Neguembor et al., 2013]. The functional role of SUV420H2 during myogenic lineage progression may suggest a regulatory role for this protein or other SUV420 homologs during lineage commitment or progression of other mesenchymal cell types, including osteoblastic cells. In this study, we present experimental evidence that SUV39H2 is the most relevant H4K20 methyl transferase during phenotype commitment in osteoblasts and is essential for osteoblast maturation.
MATERIALS AND METHODS
CELL CULTURE
MC3T3-E1, a clonal pre-osteoblastic cell line derived from newborn mouse calvaria and MLO-A5, a mature osteoblast to pre-osteocyte like cell line established from the long bones of 14-day-old mice (kindly donated by Lynda Bonewald, University of Missouri—Kansas City) were cultured in a humidified atmosphere with 5% CO2 at 37°C and were sub-cultured twice per week using Trypsin (TrypLExpress, Gibco) before achieving confluence. MC3T3-E1 and MLO-A5 cells were cultured in α-minimum essential medium (α-MEM; Gibco) containing 100 U/ml penicillin, and 100 µg/ml streptomycin (Pen-strep, Gibco); for MC3T3-E1 culture media was supplemented with 10% heat inactivated fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA) and MLO-A5 cells were cultured on rat-tail derived collagen Type I (Roche) coated dishes (final concentration of 0.15 mg/ml) and culture media was supplemented with 5% fetal calf serum (FCS, Hyclone) and 5% calf serum (CS, Hyclone).
Differentiation of MC3T3-E1 cells was induced with 50 µg/ml ascorbic acid (Sigma–Aldrich) and 5 mM β-glycerophosphate (Sigma–Aldrich) in medium containing 10% FCS. Cells were seeded in culture dishes at a density of 10,000 cells/cm2. Before cell differentiation was induced, the culture medium was changed.
Suv420H2 SIRNA TRANSFECTIONS
For Suv420h2 depletion by siRNA, MC3T3-E1 cells were seeded at 10,000 cells/cm2 in diverse multiwell plate formats for protein and RNA collection as well as for alkaline phosphatase activity measurements and for extra cellular matrix (ECM) deposition and mineralization assessment by picro sirius red staining and by alizarin red staining, respectively. Six hours after seeding, cells were transfected with scrambled or Suv420h2 specific siRNA (# L-052050-00, GE Dharmacon) using Lipofectamine RNAiMAX (Invitrogen) as described by the supplier. One day after transfection, medium change was performed and osteoblastic induction was induced as described above. Protein was isolated 3 days after transfection, RNA was isolated 3, 6, and 10 days after transfection, ECM deposition was analyzed 7 days after transfection, alkaline phosphatase activity was measured 10 days after siRNA transfection and ECM mineralization was analyzed 29 days after siRNA transfection.
PROTEIN ISOLATION AND IMMUNOBLOTTING
Whole cell protein extracts were prepared using SDS sample buffer (3% SDS and 125 mM Tris–HCl, pH = 6.8) and heated at 95°C for 5 min. For immunoblotting analysis, 5 µg of total protein extracts were separated on 11% SDS poly-acryl amide gels, transferred to polyvinylidene difluoride membranes (Millipore), blocked for 1 h with 5% non-fat dry milk in TBST buffer (137 mM NaCl, 2.7 mM KCl, 19 mM Tris, 0.1% Tween). The following primary antibodies were used: rabbit-anti-Gapdh (#5174S, Cell Signaling), mouse-anti-H4K20me3 (#39672, Active Motif), rabbit-anti-histone H4 (#61200, Active Motif), and rabbit-anti-Suv420h2 (#ab91224, Abcam). Washing was performed with TBS buffer containing 0.1% Tween. Subsequently, binding of the secondary antibody (anti-rabbit IgG/anti-mouse IgG horseradish peroxidase-coupled) (Santa Cruz) diluted 1:10,000 in 5% non-fat dry milk in TBST followed by detection with the Amersham ECL Prime Western Blot Detection Reagent (Ge Dharmacon) was carried out as described by the supplier. Chemo-luminescence was measured with an image acquisition system (Chemidoc Touch, Biorad). Measurements are given as means of three immuno-blots and representative blots are shown.
ISOLATION OF RNA AND REVERSE-TRANSCRIPTASE QUANTITATIVE POLYMERASE CHAIN REACTION (RT-qPCR)
Total RNA was extracted using the Direct-Zol RNA Mini Prep Kit (Zymo) following the supplier's instructions. cDNA was synthesized from ∼0.5 µg RNA using the SuperScript III first strand synthesis system (Invitrogen) as described by the supplier. The resulting cDNAs were subjected to quantitative PCR amplification with a real-time cycler using the QuantiTect SYBR-Green PCR Kit (Qiagen) for the genes Suv39h1, Suv39h2, Suv420h1, Suv420h2, Sp7, Bglap2, Alpl, Runx2, Ezh1, Ezh2, Ccnb2, Ibsp, Col1a1, and Gapdh (for primers, see Table I). SYBR-Green RT-qPCR was started with an initial denaturation step at 95°C for 15 min and then continued with 45 cycles consisting of 30 s denaturation at 95°C, 30 s annealing at 60°C, and 30 s extension at 72°C in a CFX384 real time system machine (Bio-Rad). All RT-qPCR assays were performed in triplicate and expression was evaluated using the comparative quantification method [Pfaffl, 2001].
| Fwd primer | Rev primer | |
|---|---|---|
| Alpl | CCAGAAAGACACCTTGACTGTGG | TCTTGTCCGTGTCGCTCACCAT |
| Bglop2 | GCAATAAGGTAGTGAACAGACTCC | CCATAGATGCGTTTGTAGGCGG |
| Ccnb2 | GCACTACCATCCTTCTCAGGTG | TGTGCTGCATGACTTCCAGGAC |
| Col1a1 | CCTCAGGGTATTGCTGGACAAC | CAGAAGGACCTTGTTTGCCAGG |
| Ezhl | TGCGTCTCAGGATGGAGGA | TTGCTATATCCATTTTCCTCATGGA |
| Ezh2 | CCTTCCATGCAACACCCAAC | CCTTAGCTCCCTCCAGATGC |
| Hist2h4 | AAGGTTCTCCGCGACAACATCC | GTCGCGGATCACATTCTCAAGG |
| Ibsp | GAATGGCCTGTGCTTTCTCG | CCGGTACTTAAAGACCCCGTT |
| Runx2 | CCTGAACTCTGCACCAAGTCCT | TCATCTGGCTCAGATAGGAGGG |
| Sp7 | GGCTTTTCTGCGGCAAGAGGTT | CGCTGATGTTTGCTCAAGTGGTC |
| Sparc | CCCCTCAGCAGACTGAAGTT | ACAGGTACCCCTGTCTCCTC |
| Suv39hl | AGGAGCGGCCAATCGAC | CAGCACACACTGCAACCTTT |
| Suv39h2 | GCCTTATTCAAGGCCTGGTG | GTCACACAAGTACTCCACCTCA |
| Suv420hl | CGGCTGCTTCCAACTCTACC | AGTGATTCCGCAGTCTGATCT |
| Suv420h2 | GCAGAGCTGCGTGAAGAGG | ACAGGCAGTATTCCCATCTGA |
GENE EXPRESSION PROFILING OF MOUSE CELLS DERIVED FROM FRACTURE CALLUS AND EZH2 CKO CALVARIA
We performed gene expression profiling by next generation RNA sequencing (RNA-seq) of calvaria bone or callus-derived cells in a mouse model for long bone healing derived from fracture callus. Calvarial bone was derived from either wild-type mice, or mice in which the Ezh2 gene was conditionally ablated using a Prrx1-Cre driver as previously described [Dudakovic et al., 2015]. Cells derived from the callus were isolated using a lineage-traced method by recombination of a floxed stop allele and activation of downstream red fluorescent protein (tomato) upon activation of a SMACreERT2 reporter [Matthews et al., 2014]. This genetic approach permits marking of all cells derived from SMACre expressing mesenchymal stem cells, which are rendered Tomato positive, while Sma-negative cells that include hematopoietic cells remain Tomato-negative. Callus cell preparations were sorted by flow cytometry to yield Tomato-positive and -negative that were subjected to RNA-seq. Raw read data from all samples were processed using a standard bioinformatic pipeline that has been described in detail previously to obtain values for mRNA levels that are expressed as reads per million mapped reads per kilobasepair.
RELATIVE QUANTIFICATION OF ECM DEPOSITION
Seven days after Suv420h2 siRNA transfection, MC3T3-E1 cells were removed using 0.5% sodium deoxycholate. The residual ECM was washed with PBS and stained with picro Sirius red as described before [Thaler et al., 2010a, 2013]. Fluorescence quantification was performed on a Tecan (Maennedorf, Switzerland) multi-well reader.
ALKALINE PHOSPHATASE ACTIVITY (ALP)
MC3T3-E1 cells were seeded in 12 multi well culture dishes at a density of 10,000/cm2 and transfected with Suv420h2 specific or scrambled siRNA as described above. On the next day, the medium was changed and cell differentiation was induced. After 10 days, cell number and ALP-activity were analyzed. For determination of cell number (DNA amount employed as surrogate), cell layers were washed with PBS and frozen with 1 mM Tris–HCl buffer (pH 8.0) containing 0.1 mM EDTA. During thawing, Hoechst 33258 dye (Polysciences, Warrington, PA) was added (1 µg/ml) and, after an incubation of 15 min at room temperature, the fluorescence was measured (excitation 360, emission 465 nm). The amount of DNA was estimated using a standard curve prepared from calf thymus DNA (Roche). Thereafter, alkaline phosphatase (ALP) activity was measured with p-nitrophenylphosphate (Sigma) (2.5 mg/ml in 0.1 M diethanolamine buffer [pH 10.5], 150 mM NaCl, 2 mM MgCl2) by incubation of the cell layers for 20 min at room temperature. Absorption was measured in a microplate reader at 405/490 nm. ALP activity per well was estimated using a standard curve prepared from calf intestinal ALP (Roche) and referred to the amount of DNA previously measured.
ALIZARIN RED STAINING
For assessment of mineralization, MC3T3-E1 cells were fixed in 4% paraformaldehyde at 29 days after siRNA transfections and 28 days after induction of osteoblastic differentiation. Fixed cells were stained with 40 mM Alizarin Red S (Sigma–Aldrich), pH 4.2, by incubating the cells for 20 min at room temperature. Subsequently, Alizarin Red S dye was extracted using 10% cetylpyridinium chloride in 10 mM sodium phosphate, pH 7.0, for 45 min at room temperature. Alizarin Red S absorbance was measured at 562 nm in a multiplate reader (Tecan) and normalized to total DNA amount which previously measured by Hoechst stain with a multiplate reader (Tecan).
STATISTICAL ANALYSIS
Statistical analyses were performed using ANOVA or Student's t-test in Prism 4.03 (GraphPad Software). Values of P ≤ 0.05 were considered significant. Each experiment consisted of at least three biological replicates. For RT-qPCR data, results from technical triplicates were averaged and the mean value was treated as a single, biological statistical unit. Results are presented as means ± SD.
RESULTS
TRANSCRIPTOME ANALYSIS BY NEXT GENERATION SEQUENCING REVEALS ELEVATED IN VIVO EXPRESSION OF SUV420H2 IN OSTEOBLASTS
We performed gene expression profiling using RNA-seq of mouse cells derived from fracture callus and calvaria. RNA-seq values were compared to assess which epigenetic regulators are most specific for mesenchymal cells and bone tissues. Heat-map analysis revealed that there are many epigenetic genes (n = 113 out of 345 total) that are selectively expressed at higher levels in mesenchymal cells or tissues with an osteo-chondrogenic biomarker signature (Fig. 1A and B). Among these genes, we noted that the six methyltransferases Ezh1, Ezh2, Suv39h1, Suv39h2, Suv420h1, and Suv420h2 are prominently expressed in callus cells and calvarial tissue (Fig. 1C).
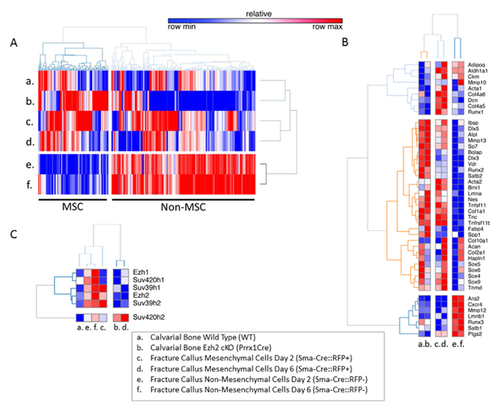
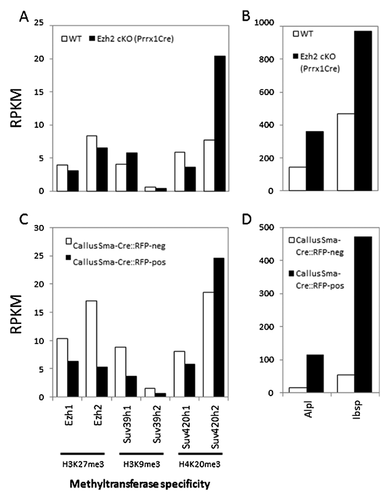
Direct comparison of the six methyltransferases in our study reveals that Suv420h2 is expressed at the highest mRNA levels compared to Ezh1, Ezh2, Suv39h1, Suv39h2, and Suv420h1 (Fig. 2A) in calvarial bone from mice with a conditional Ezh2 null mutation that accelerates osteoblast differentiation and elevates expression of the bone markers Alpl and Ibsp (Fig. 2B). Furthermore, Suv420h2 represents the only gene of this set of six that is selectively enriched in mesenchymal cells in the fracture callus at day 6 and that are marked by RFP expression by lineage-tracing (Fig. 2C). These same RFP-positive cells also express very high levels of Alpl and Ibsp compared to RFP-negative cells in the callus (Fig. 2D). Thus, the RNA-seq data indicate that Suv420h2 is selectively expressed at higher levels in tissues and cells that robustly express osteoblastic markers. This finding suggests that H4K20 methyl transferase activity may be linked to progression of osteoblast differentiation.
SUV METHYLTRANSFERASES ARE DOWN-REGULATED DURING ONSET OF QUIESCENCE AND DIFFERENTIATION IN MC3T3-E1 OSTEOBLASTS AND MLO-A5 PRE-OSTEOCYTES
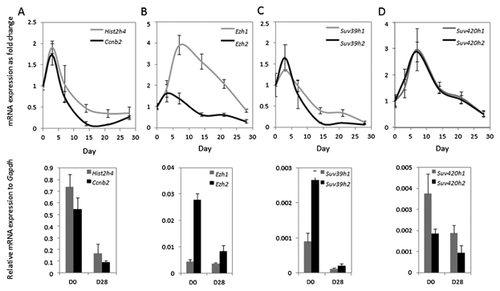
Having established that Suv420h2 is strongly expressed in osteogenic cells by RNA-seq, we validated its expression by RT-qPCR in cell culture models for osteoblast and early osteocyte differentiation (Figs. 3 and 4). As expected, the proliferation markers histone H4 (Hist2h4) and Cyclin B2 (Ccnb2) are down regulated after day 3 in both MC3T3-E1 and MLO-A5 cells (Figs. 3A and 4A). Similarly, Ezh2, Suv39h1, and Suv39h2 also have peak expression during the early culturing period in MC3T3-E1 cells (Fig. 3B and C). However, Ezh1, Suv420h1, and Suv420h2 appear to be expressed at maximal levels at day 7 and their expression gradually fades during progression of osteoblast differentiation over 28 days of MC3T3-E1 cell culture (Fig. 3B–D). For comparison, expression patterns in MLO-A5 osteocytes tend to be generally similar, albeit more variable and expression changes are less pronounced (Fig. 4B and D). We note that Suv420h2 is expressed at lower levels and shows less modulation in its expression compared to Suv420h1 in MLO-A5 osteocytes (Fig. 4D). Taken together, these data corroborate the in vivo expression data presented in Figure 1 and indicate that Suv420h2 is actively expressed during osteoblast maturation but is expressed at lower levels after the osteoblast-osteocyte transition.

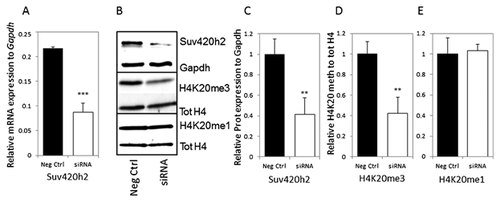
SUV420H2 CONTROLS H4K20 TRI-METHYLATION IN PRE-OSTEOBLASTIC CELLS
H4K20 tri-methylation is a hallmark of transcriptional repression and is regulated by Suv420h1 and Suv420h2. Because Suv420h2 is robustly expressed in osteoblastic cells (see Figs. 1-4), we analyzed the effects of Suv420h2 knock down by siRNA in MC3T3-E1 pre-osteoblasts to understand its functional role in osteoblastogenesis. We observe a clear two- to threefold reduction in Suv420h2 expression at both RNA (Fig. 5A) and protein levels (Fig. 5B and C) at 3 days after siRNA transfection. More importantly, immunoblot analysis shows that depletion of Suv420h2 reduces H4K20me3 levels (Fig. 5D) but not H4K20me1 levels (Fig. 5E). These studies support the conclusion that Suv420h2 is rate-limiting for global H4K20me3 levels in MC3T3-E1 cells and thus a major regulator of H4K20me3-directed transcriptional repression during osteoblastogenesis.
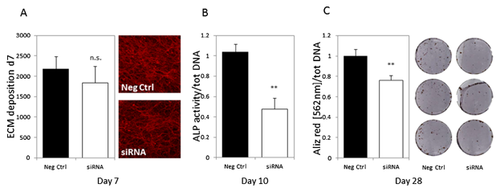
PRIMARY OSTEOGENIC FUNCTIONS ARE DISRUPTED AFTER REDUCTION OF SUV420H2 EXPRESSION
Extra cellular matrix (ECM) deposition and its mineralization are among the main activities of differentiating osteoblasts. To elucidate the role of Suv420h2 on ECM maturation during osteoblastogenesis, we analyzed the effect of Suv420h2 depletion by siRNA on ECM deposition around MC3T3-E1 osteoblasts. Collagen fibers were visualized using Picro Sirius Red and staining intensity was determined at day 7 after initiation of osteoblastic differentiation. We observed a modest albeit not significant reduction in ECM deposition in MC3T3-E1 cells treated with Suv420h2 siRNAs (Fig. 6A). However, alkaline phosphatase activity, an enzyme marker that is deposited in the collagen-rich ECM during osteoblast maturation and ECM mineralization, was significantly decreased at day 10 in cells in which Suv420h2 levels were reduced by siRNA treatment (Fig. 6B). Furthermore, ECM mineralization was significantly reduced by about 25% in MC3T3-E1 cells at day 28 of differentiation (Fig. 6C), even though Suv420h2 expression was only actively suppressed by siRNA during the initial stages of the culture period. The lasting effects of a temporary knock down of Suv420h2 by siRNA indicates that Suv420h2 may affect the molecular memory of MC3T3-E1 cells through epigenetic mechanisms operative at early stages of osteoblastogenesis.
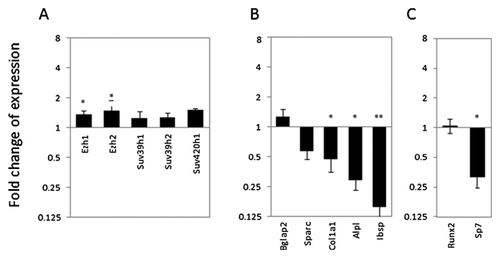
SIRNA INHIBITION OF Suv420H2 NEGATIVELY AFFECTS THE EXPRESSION OF HISTONE METHYLTRANSFERASES AND OF PRIMARY OSTEOBLASTIC MARKERS IN DIFFERENTIATING MC3T3-E1 CELLS
Because the long-term effects of Suv420h2 inhibition on osteoblastic differentiation of MC3T3-E1 cells are instigated during the first several days of the culturing period, we analyzed the acute regulatory functions of Suv420h2 by analyzing gene expression of multiple markers and select transcription factors genes in cells in which Suv420h2 expression was reduced by siRNA (see Fig. 5). Our analysis focused on day 3 of MC3T3-E1 differentiation (Fig. 7) as a key time-point within a broader time-course up to day 10 (Figs. 7-9). Depletion of Suv420h2 does have small (less than 1.5-fold), although partially significant effects on the expression of other methyltransferases (Fig. 7A). However, cells exhibit two- to eightfold a down-regulation of several ECM-related osteoblastic genes, including Sparc, Col1a1, Alpl, and Ibsp (Fig. 7B). We did not detect significant changes in the levels of Bglap2, presumably because cells at day 3 in culture are immature and do not have a robust basal level of expression. More importantly, we observed a strong reduction in the expression of the osteoblastic transcription factor Sp7 (Osx) at day 3 after Suv420h2 siRNA transfection, while Runx2 levels remain relatively constant (Fig. 7C). These results suggest that Suv420h2 alters key osteoblastic markers at early stages of osteoblastic differentiation in cell culture.
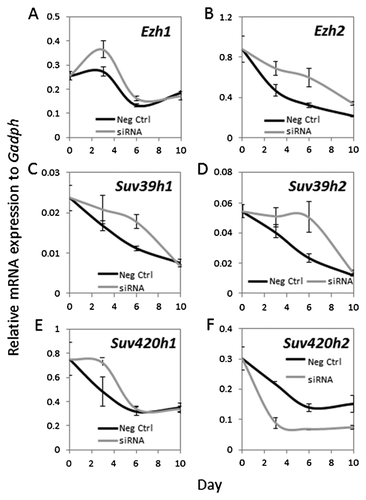
LONG-TERM EFFECTS OF TRANSIENT Suv420H2 INHIBITION ON THE EXPRESSION OF PRIMARY OSTEOBLASTIC MARKERS IN DIFFERENTIATING MC3T3-E1 CELLS
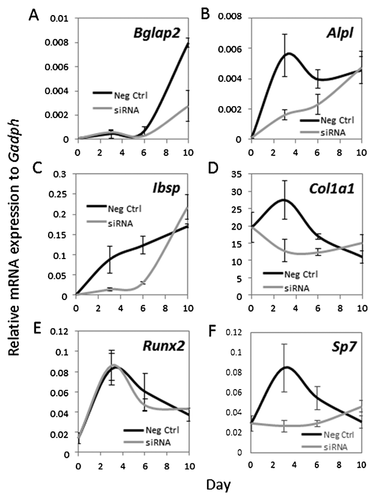
Transient depletion of Suv420h2 expression by siRNA for the first 3 days at early stages of osteoblast differentiation affects late osteoblastic events like ECM mineralization (see Fig. 6C). Therefore, we assessed whether these early reductions in Suv420h2 levels have sustained effects on osteoblastic maturation (Figs. 8 and 9). Interestingly, although MC3T3-E1 cells were treated with Suv420h2 siRNA for only 3 days, Suv420h2 expression remains low throughout the remainder of the time course until day 10 (Fig. 8F). Even though this reduced level of Suv420h2 has only modest effects on the expression of other epigenetic regulators at day 3 (Fig. 8A–F; see also Fig. 7A), greater changes in expression of Ezh2, Suv39h1, and Suv39h2 are observed at day 6 (Fig. 8B–D). Thus, loss of Suv420h2 expression may have broad ranging secondary epigenetic effects on gene expression during osteoblast differentiation (Fig. 8F). Furthermore, transient Suv420h2 knock down (Fig. 8F) has primarily transient effects on most osteoblastic markers like Col1a1, Ibsp, Alpl (Fig. 9B–D) or osteoblastic transcription factors like Sp7 (Fig. 9E and F). However, Suv420h2 siRNA treatment has a delayed effect on the late-stage osteoblast marker Bglap2 at day 10 (Fig. 9A). We conclude that transient knock down of Suv420h2 has delayed effects on ECM mineralization by altering lysine-methylation related epigenetic mechanisms, and these alterations reduce expression of early osteoblastic markers and delay expression of late osteoblast markers that together control ECM maturation.
DISCUSSION
Epigenetic control of gene expression by histone modifications is a principal regulatory mechanism by which cells control lineage commitment and cell maturation. In this manuscript, we present key evidence for the important role of H4K20 methylation and the related histone methyl transferase Suv420h2 in osteoblastic differentiation. RNA-seq profiling revealed that Suv420h2 expression is elevated in cells and tissues which robustly express osteoblastic markers. Consistent with its functional role in bone cell differentiation, Suv420h2 depletion by siRNA suppresses the maturation potential of MC3T3-E1 pre-osteoblastic cells by reducing Alp activity, ECM mineralization, as well as the expression of osteoblast-related biomarkers (e.g., Ibsp, Col1a1, or Sp7). Our finding that Suv420h2 is a positive regulator of osteoblast differentiation complements our previous studies that established the importance of other epigenetic regulators, including the H3K27 methyl transferase EZH2 [Dudakovic et al., 2015, 2016], DNA hydroxymethylases TET1 and TET2 [Thaler et al., 2012, 2016], DNA methyl-transferases [Thaler et al., 2010b, 2011] and histone deacetylases [Dudakovic et al., 2013]. Interestingly, each of these epigenetic regulators are enzymes whose activity can be modulated by natural or synthetic inhibitors that strongly affect the differentiation potential of osteoblasts [Thaler et al., 2012, 2016; Fu et al., 2013; St John et al., 2014]. Taken together, our studies corroborate general concepts in the field that the epigenome of osteoblasts is critical for responses to osteogenic signaling pathways and ligands (e.g., WNT, BMP2, Vitamin D3) [Stein et al., 2010; Wu et al., 2014; Gordon et al., 2015; Lee et al., 2015; Meyer et al., 2016].
We observed that short-term depletion of Suv420h2 expression by siRNA treatment decreases H4K20 methylation, and immediately disrupts the natural temporal expression of early osteoblast-specific genes including Alpl, Sp7, Col1a1, Ibsp, and Sparc, as well as that of the late osteoblastic marker Bglap2. These disruptions in the temporal sequence of bone specific gene expression clarify the observed inhibition of ECM mineralization upon disruption of Suv420h2-mediated H4K20 methylation. Importantly, we observed these biological effects when osteoblastic cells treated with siRNA for 1 day prior to the induction of osteoblast differentiation during a time course that lasts up to 28 days. Thus, Suv420h2 might play a role in the epigenetic programming of specific osteoblastic pathways at early differentiation stages. The attenuation in ECM mineralization indicates that epigenetic priming of osteoblastic cells at early osteoblastic differentiation stages has relatively long term consequences.
The biological importance and molecular significance of H4K20 methylation by Suv420h2 has not yet been fully resolved. For example, mono-methylation of H4K20 is related to transcriptional activation [Beck et al., 2012]. However, H4K20me2 and H4K20me3 histone marks have been reported to be important for silencing gene expression through formation of heterochromatic regions [Schotta et al., 2004]. Our studies which show that Suv420h2 is a key epigenetic regulator in osteoblasts will provide a strong impetus for future studies aimed at defining the specific role of H4K20 mono-, di-, and tri-methylation during osteoblast differentiation, skeletal development, and bone homeostasis in vivo.
ACKNOWLEDGMENTS
We thank our colleagues at Mayo Clinic, and especially Jennifer Westendorf for support and stimulating discussions. We also thank Dr. Lynda Bonewald (University of Missouri-Kansas City, Kansas, MI) for generously providing cell lines. We also appreciate the assistance of Ivo Kalajzic and for providing sorted lineage-traced cells derived from a mouse fracture healing model.
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases [R01 AR049069 to AJVW; F32 AR066508 to AD] and a fellowship grant from the Center of Regenerative Medicine at Mayo Clinic (to RT) as well as by the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) grant number: 1130931 (to MG), and Iniciativa Científica Milenio. Ministerio de Economía, Fomento y Turismo grant number P09/016!F (to MG). We also appreciate the generous philanthropic support of William H. and Karen J. Eby, and the charitable foundation in their names.