Detection of FLT3/TKD and IDH1 Mutations in Pakistani Acute Myeloid Leukemia Patients by Denaturing HPLC
ABSTRACT
Acute myeloid leukemia (AML) is characterized by an increase in the number of myeloid cells in the marrow and an arrest in their maturation. Various genetic mutations are associated with AML. FMS-like tyrosine kinase 3 (FLT3), a member of the class III receptor tyrosine kinase family, plays an important role in stem cell survival, and the development of dendritic and natural killer cells. FLT3/TKD mutations are generally missense mutations or in-frame alterations of residues D835 and I836 within the activation loop of the FLT3 protein. D835 mutations have been reported to occur in ≈ 7% of AML patients. Mutations have also been reported in exon 4 of isocitrate dehydrogenase 1 (IDH1) in ≈9% of AML patients. Mutations in FLT3/TKD and IDH1 genes were studied in AML patients from Pakistan and correlated with the laboratory findings. FLT3/TKD mutations were found in 7%, while IDH1 mutations were found in 10% Pakistani AML patients. Neither of these mutations was significantly correlated with age and sex, although the incidence of these mutations was higher in female patients. These mutations were found to be positively associated with each other. IDH1 mutations were positively associated with FAB type M1 and negatively associated with FAB type M2. In conclusion, the overall incidence of all these mutations in Pakistani patients was within the globally reported ranges. J. Cell. Biochem. 118: 1174–1181, 2017. © 2016 Wiley Periodicals, Inc.
Acute myeloid leukemia (AML) is characterized by an increase in the number of myeloid cells in the marrow and an arrest in their maturation, frequently resulting in hematopoietic insufficiency (granulocytopenia, thrombocytopenia, or anemia), with or without leukocytosis.
FMS-like tyrosine kinase 3 (FLT3), also known as stem cell tyrosine kinase-1 (STK1) or fetal liver tyrosine kinase-2 (FLK-2), is a member of class III tyrosine kinase (TK) receptors. It includes five immunoglobulin-like loops in the extracellular region, a transmembrane region, an intracellular juxta-membrane (JM) domain, two tyrosine kinase (TK) domains interrupted by a kinase insert, and a C-terminal tail [Agnes et al., 1994]. The gene is located at chromosome 13q12 and consists of 24 exons [Rosnet et al., 1991; Abu-Duhier et al., 2001a].
The expression of FLT3 is found mainly in hematopoietic stem cells in the bone marrow, lymph nodes, and thymus [Rosnet et al., 1993], but is also seen in other tissues including cerebellum, placenta, gonads, and brain [Maroc et al., 1993]. After the ligand binds, the receptor dimerizes, and autophosphorylates, which activates downstream signaling pathways, resulting in the proliferation of pluripotent stem cells [Lyman, 1995]. Other cytokines such as Kit ligand also influence this interaction. In fact, FLT3 ligand or KIT ligand alone induce very little proliferation of in vitro human progenitor cells. But, when used together, they have a synergistic effect on stem cell proliferation [Hannum et al., 1994].
The FLT3 gene is expressed in 93% of AML, 100% of B cell ALL, and 87% of T-cell ALL patients [Drexler, 1996]. In AML, over expression of FLT3 mRNA has been reported in many studies, suggesting that this high level of expression may have some critical role in increased proliferation and survival of the leukemic cells [Carow et al., 1996]. Two types of mutations have been reported in the FLT3 gene in AML patients viz., internal tandem duplications (FLT3/ITD) and mutations in the second tyrosine kinase domain (FLT3/TKD). FLT3/ITDs result from the duplication and tandem insertion of a portion of the juxtamembrane (JM) region. The duplicated region is always present between exons 14 and 15 of the FLT3 gene [Nakao et al., 1996]. The FLT3/ITDs are always in frame and produce functional FLT3 protein. FLT3/ITDs are one of the most frequent mutations reported in AML, found in 13–32% of adult AML patients [Gale, 2003], and are associated with worse prognosis. FLT3/TKD mutations in exon 20 include at least six different substitutions within the D835 codon, predominantly tyrosine and histidine, less frequently valine, glutamate, and asparagine. The deletion of I836 and insertion of nucleotides have also been detected, but the sequence always remains in frame. The incidence of these mutations varies between 5% and 7% in adult AML patients [Abu-Duhier et al., 2001b; Yamamoto et al., 2001]. The impact of FLT3/TKD mutations on prognosis is unclear.
Isocitrate dehydrogenase (IDH) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate, and carbon dioxide in the citric acid cycle. Three IDHs have been reported: one NAD(+)-dependent IDH, which is cytosolic, and two NADP(+)-dependent IDHs, one of which is mitochondrial and the other cytosolic. Each NADP(+)-dependent isozyme is a homodimer. The IDH1 gene is on chromosome 2 at 2q33.3 and consists of 30.12 kb. It encodes one of the NADP(+)- dependent isocitrate dehydrogenases, which is found in the cytoplasm and peroxisomes. It plays a significant role in cytoplasmic NADPH production.
Point mutations in the exon 4 (R132) of the IDH1 gene were first reported in 12% cases of glioma [Parsons et al., 2008]. Similar IDH1 mutations were also found in more than 70% of the WHO grade I and III astrocytomas, and in glioblastomas. These mutations have also been identified in 9% of primary AML patients, and are more associated with normal karyotype AML (16%) [Mardis et al., 2009]. In a study on de novo AML adult patients, 10% patients with normal karyotype were found to have IDH1 R132 mutations. These mutations were found to be associated with isolated trisomy 8, NPM1 mutations, and FAB subtype M1 [Chou et al., 2010].
In the present study, FLT3/TKD and IDH1 mutations were studied in 70 AML patients from Pakistan and correlated with hematological findings.
MATERIALS AND METHODS
SUBJECTS
Samples from 70 Pakistani AML patients were included in this study. The AML patients were selected according to standard hematological and clinical parameters with the help of consultant hematologists at different hospitals in Lahore, Pakistan from January 2006 to February 2009. Clinical and laboratory findings were obtained on a prescribed proforma from the hospital. DNA was extracted from all the samples using the Guanidine Thiocyanate and ethanol precipitation method [Malferrari et al., 2002]. DNA concentrations were measured on a NanoDrop ND-1000 spectrophotometer and each DNA sample was diluted to a final working concentration of 30 ng/μl with TE buffer for further use in PCR.
PCR AMPLIFICATION
For the screening of FLT3/TKD mutations, PCR was used to amplify a 278 base pair fragment covering FLT3 exon 20. For IDH1 mutations screening, a 369 base pair fragment covering IDH1 exon 4 was amplified. For both FLT3/TKD and IDH1, the PCR mixture contained 1× Optimase buffer, 1.5 mM MgSO4, 0.2 mM dNTPs, 0.5 mM each primer (17F2 and 17R3 for FLT3/TKD, 4F and 4R for IDH1, Table I), 0.5 U Optimase polymerase (Transgenomic Limited, Glasgow, UK), and 30 ng DNA in a total reaction volume of 20 μl. Cycling conditions for PCR were: Initial denaturation at 95°C for 5 min and then 35 cycles each of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. A known wild type and mutant-positive sample were also amplified along with patient samples. Five micro liters of each PCR product were run on a 2% agarose gel in 1× TBE buffer to confirm the presence of PCR product. To enhance the formation of heteroduplexes before running on DHPLC, the PCR products were denatured at 95°C for 4 min and then allowed to cool down slowly to room temperature at a rate of 1°C/min.
| Gene/exon | Primer sequence | WT product size (bp) |
|---|---|---|
| FLT3/20 | 17F2: 5′-CATCACCGGTACCTCCTACTG-3′ | 278 |
| 17R3: 5′-TAACGACACAACACAAAATAGCCGT-3′ | ||
| IDH1/4 | 4F: 5′-CATTTGTCTGAAAAACTTTGCTTC-3′ | 369 |
| 4R: 5′-ACATGCAAAATCACATTATTGCCA-3′ | ||
| IDH1/4 | 4F2: 5′-ATCACTCCTGATGAGAAGAGGGTTGAG-3′ | 200 |
| R132H/MM(R): 5′-ACATGACTTACTTGATCCCCATAAGCATCA-3′ | ||
| R132C/MM(R): 5′-CATGACTTACTTGATCCCCATAAGCATGTC-3′ | 199 |
DHPLC RUNNING CONDITIONS
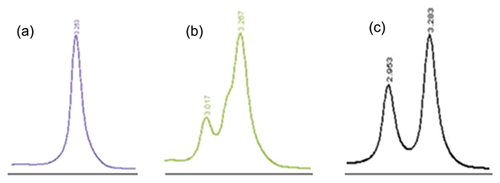
Flow rate of mobile phase was 0.9 ml/min. The gradient of buffer A and B contained 55% buffer B at the beginning with an increase of 2% per minute to 63% at the end. The duration of the gradient was 4 min. The temperature of the column oven was 59.0°C for FLT3/TKD and 57.0°C and 59.0°C for IDH1. Volume of PCR product per injection was 5 μl. The ultraviolet detector was set at 260 nm. Heterozygous profiles were identified visually on the basis of the appearance of additional earlier peaks of eluting DNA on the chromatograms. Homozygous samples showed only one peak. The relative mutant level was visually assessed using the height of the mutant peak compared with that of wild type peak of the same sample run.
CONFIRMATION OF FLT3/TKD MUTATIONS BY ECORV DIGESTION AND SEQUENCING
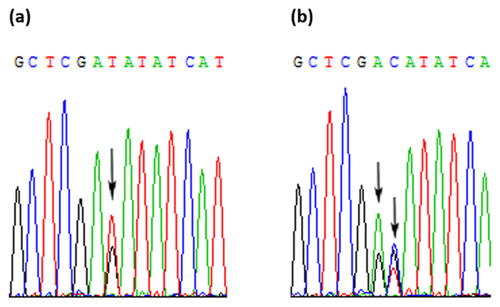
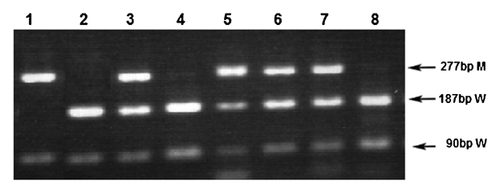
The samples which showed an additional peak on the DHPLC chromatogram were PCR amplified and digested with EcoRV. The PCR mixture contained 1× Bioline buffer, 1.0 mM MgCl2, 0.2 mM dNTPs, 0.5 mM each primer (17F2 and 17R3), 0.5 U Taq polymerase (Bioline, London, UK), and 30 ng DNA. The total reaction volume was 20 μl. Cycling conditions for PCR were: 35 cycles each of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. A known wild type and mutant-positive sample were also amplified along with unknown samples. The PCR products were run on a 2% agarose gel. Five micro liters of each PCR product were digested with EcoRV (New England BioLabs, Hitchin, UK). The digestion mixture contained 20 U EcoRV, 1× NEB buffer 3, and 1 μg BSA. The final volume was adjusted to 10 μl and placed at 37°C for 4–5 hr. The digested products were run on a 2% agarose gel. The wild type fragments were digested into two fragments of 187 and 91 bp in size while the mutant products remained uncut at 278 bp. The PCR products generated using Bioline reagents were cleaned up using QIAquick PCR purification kit as per manufacturer's protocol and were sent to an in-house sequencing service.
CONFIRMATION OF IDH1 P.R132 MUTATIONS BY RESTRICTION DIGESTION
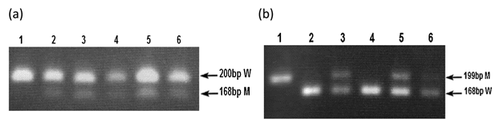
For R132H mutations, amplicons of 200 bp were generated using Bioline PCR with primers IDH1ex4/F2 and IDH1/R132H/MM(R) (Table I). The PCR mixture contained 1× Bioline buffer, 1.0 mM MgCl2, 0.2 mM dNTPs, 0.5 mM each primer, 0.5 U Taq polymerase (Bioline, London, UK) and 30 ng DNA. The total reaction volume was 20 μl. Cycling conditions for PCR were: 35 cycles each of 95°C for 30 s, 64°C for 30 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. PCR products were digested with BspHI (New England BioLabs, Hitchin, UK) using manufacturer's recommendations. The digested products were run on a 2% agarose gel. Wild-type (WT) amplicons were uncut (200 bp) and R132H-positive amplicons were reduced to 168 bp.
For R132C, R132S, and R132L mutations, amplicons of 199 bp were generated using Bioline PCR with primers IDH1ex4/F2 and IDH1/R132C/MM(R) (Table I). The PCR mixture contained 1X Bioline buffer, 1.0 mM MgCl2, 0.2 mM dNTPs, 0.5 mM each primer, 0.5 U Taq polymerase (Bioline, London, UK), and 30 ng DNA. The total reaction volume was 20 μl. Cycling conditions for PCR were: 35 cycles each of 95°C for 30 s, 64°C for 30 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. PCR products were digested with TaqI (New England BioLabs, Hitchin, UK) using manufacturer's recommendations. The digested products were run on a 2% agarose gel. WT amplicons were reduced to 168 bp but amplicons carrying an R132C, R132S, or R132S mutation remained uncut (199 bp). All the IDH1 mutations were also confirmed by sequencing.
RESULTS
AML PATIENTS
Out of these 70 patients, 38 were male, and 32 were female. Median values and range of age, WBC count, platelet count, and %blasts for males and females along with FAB types are shown in Table II.
| Total (n = 70) | Male (n = 38) | Female (n = 32) | |
|---|---|---|---|
| Age (years) | |||
| Median | 30 | 32 | 24 |
| Range | 9–68 | 16–68 | 9–50 |
| WBC count (×109 per litre) | |||
| Median | 26 | 24 | 32 |
| Range | 0.8–196 | 0.8–141 | 1.2–196 |
| Platelet count (×109 per litre) | |||
| Median | 64 | 53 | 76 |
| Range | 7–322 | 7–187 | 12–322 |
| % Blasts | |||
| Median | 42 | 48 | 41.5 |
| Range | 15–98 | 15–98 | 18–82 |
| FAB types | |||
| M0 | 0 | 0 | 0 |
| M1 | 18 | 11 | 7 |
| M2 | 24 | 13 | 11 |
| M3 | 4 | 2 | 2 |
| M4 | 9 | 5 | 4 |
| M5 | 7 | 2 | 5 |
| M6 | 3 | 2 | 1 |
| M7 | 0 | 0 | 0 |
| Unknown | 5 | 3 | 2 |
There was no difference in the median values for WBC count (P = 0.38), platelet count (P = 0.26) and % blasts (P = 0.13) of the male and female patients. Median age of male patients was significantly higher than that of female patients (P = 0.03). There was no patient with either M0 or M7 FAB type. FAB type M2 had the maximum number of patients followed by M1 and M4. Relatively smaller number of patients was seen in M3, M5, and M6 (Table II).
FREQUENCY OF MUTATIONS
FLT3/TKD mutations were detected in five patients (7%) while IDH1 mutations were detected in seven patients (10%). The incidence of FLT3/TKD mutation was almost equal among male (8%) and female (6%) patients (P = 0.95) while that of IDH1 mutations was higher in male (13%) than female (6%) patients but this was not statistically significant (P = 0.34, Table III). The overall incidence of both the mutations was slightly higher in male (21%) than in the female (12.5%) patients but this was not significant (P = 0.42).
| FLT3/TKD | IDH1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | WT | MT (%) | P | % MT | WT | MT (%) | P | % MT | |
| Sex | |||||||||
| Male | 38 | 35 | 3 (60) | 0.79 | 8 | 33 | 5 (71) | 0.34 | 13 |
| Female | 32 | 30 | 2 (40) | 6 | 30 | 2 (29) | 6 | ||
| Age (years) | |||||||||
| <25 | 30 | 27 (41) | 3 (60) | 0.66 | 10 | 28 (44) | 2 (29) | 0.02 | 7 |
| 26–50 | 36 | 34 (52) | 2 (40) | 6 | 33 (52) | 3 (43) | 8 | ||
| >50 | 4 | 4 (6) | 0 (0) | 0 | 2 (3) | 2 (29) | 50 | ||
| FAB types | |||||||||
| M1 | 18 | 17 (26) | 1 (20) | 0.76 | 6 | 15 (24) | 3 (43) | 0.27 | 17 |
| M2 | 24 | 22 (34) | 2 (40) | 0.78 | 8 | 22 (35) | 2 (29) | 0.74 | 8 |
| M3 | 4 | 4 (6) | 0 (0) | 0.43 | 0 | 4 (6) | 0 (0) | 0.35 | 0 |
| M4 | 9 | 8 (12) | 1 (20) | 0.62 | 11 | 9 (14) | 0 (0) | 0.15 | 0 |
| M5 | 7 | 7 (11) | 0 (0) | 0.29 | 0 | 6 (10) | 1 (14) | 0.69 | 14 |
| M6 | 3 | 3 (5) | 0 (0) | 0.50 | 0 | 3 (5) | 0 (0) | 0.42 | 0 |
| Unknown | 5 | 4 (6) | 1 (20) | 20 | 4 (6) | 1 (14) | 20 | ||
| WBC count ×109/L | |||||||||
| <10 | 24 | 23 (35) | 1 (20) | 0.87 | 4 | 24 (38) | 0 (0) | 0.22 | 0 |
| 10–50 | 19 | 17 (26) | 2 (40) | 11 | 17 (27) | 2 (29) | 11 | ||
| 51–100 | 13 | 12 (18) | 1 (20) | 8 | 10 (16) | 3 (43) | 23 | ||
| >100 | 9 | 8 (12) | 1 (20) | 11 | 8 (13) | 1 (14) | 11 | ||
| Unknown | 5 | 5 (8) | 0 (0) | 0 | 4 (6) | 1 (14) | 20 | ||
| Platelet count ×109/L | |||||||||
| 01–50 | 25 | 23 (35) | 2 (40) | 0.25 | 8 | 24 (38) | 1 (14) | 0.37 | 4 |
| 51–100 | 21 | 18 (28) | 3 (60) | 14 | 19 (30) | 2 (29) | 10 | ||
| >100 | 18 | 18 (28) | 0 (0) | 0 | 15 (24) | 3 (43) | 17 | ||
| Unknown | 6 | 6 (9) | 0 (0) | 0 | 5 (8) | 1 (14) | 17 | ||
| % Blasts | |||||||||
| 01–50 | 39 | 36 (55) | 3 (60) | 0.81 | 8 | 37 (59) | 2 (29) | 0.29 | 5 |
| 51–100 | 26 | 24 (37) | 2 (40) | 8 | 22 (35) | 4 (57) | 15 | ||
| Unknown | 5 | 5 (8) | 0 (0) | 0 | 4 (6) | 1 (14) | 20 | ||
FLT3/TKD mutations were found to be equally distributed among all age groups (P = 0.66). A significantly higher incidence of the IDH1 mutation was seen in elderly patients (>50 years of age, P = 0.02). Neither of the mutations was significantly associated with sex, any specific FAB type, WBC count, platelet count, or blast percentage.
FLT3/TKD MUTATIONS
In the five FLT3/TKD mutant-positive cases, D835Y was the most common FLT3/TKD mutation, found in four samples as a sole mutation, accounting for 80% of the mutant-positive patients, and 6% of the total cohort. The fifth patient had three different mutations with different levels; D835H being high, R834R intermediate, and D835Y being low (Figs. 1 and 2). The restriction digestion results confirmed the DHPLC results (Figs. 1 and 3).
IDH1 MUTATIONS
In the seven IDH1 mutant-positive cases, three different types of mutations were detected. R132C and R132H were detected at the same frequency (three in seven, 43%). R132S mutation was seen in one case. The DHPLC results (Fig. 4) were confirmed by restriction digestion (Fig. 5) and sequencing (Fig. 6).
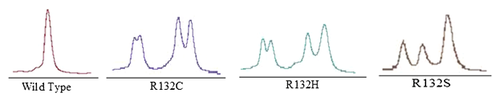
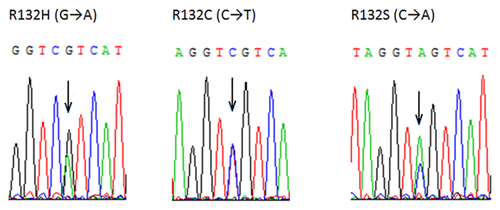
DISCUSSION
In this study, the mutations in FLT3/TKD and IDH1 (exon 4) genes were studied in AML patients of Pakistan. The association of these mutations with each other and with clinical characteristics of the patients was also studied.
The male to female ratio in Pakistani AML patients was 1.2:1.0 (56% male, 45% female) which was in accordance with US National Cancer Institute statistics about Asian populations.
FLT3/TKD MUTATIONS
Developing appropriate molecular biology techniques to detect different molecular abnormalities in human malignancies is a major challenge. Even in AML alone, there are many different reported genetic mutations. Information derived from the detection of some of these mutations is important for clinical management of the disease. Low level mutations can be very difficult to detect using conventional mutation detection techniques. It is necessary therefore to use sensitive and high throughput techniques that are capable of detecting these mutations.
Using DHPLC, FLT3/TKD mutations were detected in 7% of the Pakistani cohort. There are a number of advantages in using DHPLC over the standard EcoRV restriction enzyme digestion method that has been used in the majority of previous studies. Firstly, it is more sensitive. In the current study, there was one case which was EcoRV negative but DHPLC positive. The EcoRV-digested PCR product was therefore used as the template in a fresh FLT3/TKD PCR. When the product was digested with EcoRV, a mutant band was prominent on the gel. This suggests that the mutation level in that patient was very low. In addition, interpretation of EcoRV digestion greatly depends on the efficiency of the restriction digest. In some cases, it was extremely difficult to differentiate between an undigested low level mutant and the incomplete digestion of FLT3/WT. Using DHPLC however, such low level mutants were detected efficiently. This observation highlights the sensitivity of the DHPLC technique for the detection of mutations that are present in a small proportion of the amplified alleles. Another major limitation of restriction digestion screening is that it detects mutations only within a very specific region of DNA, usually 4–6 base pairs. Although the most frequent FLT3/TKD mutations occur within codons 835 and 836 and disrupt the EcoRV digestion site, a number of mutations occur outside this region. The DHPLC technique, on the other hand, detects mutations across a wider region of DNA. Furthermore, the chromatogram profile produced by different mutations was found to be highly specific and reproducible, in most cases enabling direct mutation identification from the WAVE pattern without the need for sequencing (Figs. 1 and 4). Considering the clinical management of AML patients with FLT3/TKD mutations, there are clear advantages to the use of DHPLC.
In the current cohort, a D835 mutation was seen in four cases as a sole mutation. Interestingly, one of the mutant positive patients had three different mutations; D835H, D835Y, and R834R. Presence of more than one mutation has already been reported [Yamamoto et al., 2001]. The EcoRV digested PCR product was used as a template for re-amplification of the PCR product for sequencing in that case.
FLT3/TKD mutations were not associated with age or sex. This was in accordance with other studies [Yamamoto et al., 2001; Mead et al., 2007]. The mutations were not found to be associated with any of the FAB types. Some studies have shown a positive correlation of the mutations with FAB type M5 [Thiede et al., 2002; Bacher et al., 2008] and a negative correlation with FAB types M2 and M6 (8) while others have shown no association with any of the FAB types [Mead et al., 2007; Whitman et al., 2008].
IDH1 MUTATIONS
Acquired mutations in the IDH1 gene have been identified in 8% [Mardis et al., 2009] and 5.5% [Chou et al., 2010] of newly diagnosed AML cases. In the current study, IDH1 mutations were seen in 10% (Pakistani samples) of AML patients. R132C and R132H compete for being the most frequent IDH1 mutations in AML [Mardis et al., 2009; Abbas et al., 2010]. In present study, R132C and R132H were detected at the same frequency (3 in 7, 43%). The mutations were found to be positively associated with higher age (P = 0.02, Table III). No association of the mutations with sex or FAB types was seen.
The overall frequency of the FLT3/TKD and IDH1 mutations in this study was higher than most of the other studies. The most likely reason for this is the use of DHPLC, which is a more sensitive technique than other conventional mutation detection techniques.
COINCIDENCE OF FLT3/TKD AND IDH1 MUTATIONS WITH OTHER DESCRIBED MUTATIONS
Using DHPLC, we have previously reported the frequency of NPM1 mutations and FLT3/ITD mutations in the Pakistani AML patients [Ali et al., 2013a,2013b]. Table IV shows the coincidence of FLT3/TKD mutations with the previously described FLT3/ITD and NPM1 mutations. The percentage of FLT3/TKD mutations in FLT3/ITD and NPM1 mutant-positive patients was not much different from that of the wild type patients. No FLT3/TKD mutation was seen in IDH1 mutant-positive patients. No significant association was seen between FLT3/TKD mutation and any of the other three mutations.
| FLT3/TKD WT (% in total WT) | FLT3/TKD mutant (% in total mutants) | P | % FLT3/TKD mutant | |
|---|---|---|---|---|
| Total (100) | 93 | 7 | – | 7 |
| FLT3/ITD wild type | 77 (83) | 6 (86) | 0.84 | 7 |
| FLT3/ITD mutant | 16 (17) | 1 | 6 | |
| NPM1 wild type | 73 (78) | 5 (71) | 0.66 | 6 |
| NPM1 mutant | 20 (22) | 2 (29) | 10 | |
| IDH1 wild type | 58 (89) | 5 | 0.29 | 8 |
| IDH1 mutant | 7 (11) | 0 (0) | 0 |
Table V shows the coincidence of IDH1 mutations with previously reported FLT3/ITD and NPM1 mutations in the acute myeloid leukemia patients of Pakistan. Although no IDH1 mutation was seen in FLT3/ITD mutant-positive patients, this was not statistically significant (P = 0.11). Same was the case with FLT3/TKD mutant-positive patients (P = 0.29). Incidence of the IDH1 mutations in mutant-positive patients was almost equal to that of the wild type patients (P = 0.83). So, IDH1 mutations were not found to be significantly associated with any of the three mutations.
| IDH1 WT (%) | IDH1 mutant (%) | P | % mutant | |
|---|---|---|---|---|
| Total (70) | 63 | 7 | 10 | |
| FLT3/ITD wild type | 52 (83) | 7 (100) | 0.11 | 12 |
| FLT3/ITD mutant | 11 (17) | 0(0) | 0 | |
| FLT3/TKD wild type | 58 (92) | 7 (100) | 0.29 | 11 |
| FLT3/TKD mutant | 5 (8) | 0 (0) | 0 | |
| NPM1 wild type | 52 (83) | 6 (86) | 0.83 | 10 |
| NPM1 mutant | 11 (17) | 1 (14) | 8 |
CONCLUSIONS
Mutations in FLT3/TKD and IDH1 genes were studied in AML patients from Pakistan and correlated with the laboratory findings. FLT3/TKD mutations were found in 7%, while IDH1 mutations were found in 10% Pakistani AML patients. Neither of these HPLC mutations was significantly correlated with age and sex, although the incidence of these mutations was higher in female patients. These mutations were found to be positively associated with each other. IDH1 mutations were positively associated with FAB type M1 and negatively associated with FAB type M2. In conclusion, the overall incidence of all these mutations in Pakistani patients was within the globally reported ranges.
ACKNOWLEDGMENT
The financial support of Higher Education Commission of Pakistan in the form of a Fellowship to the first author under the indigenous Ph.D. program is gratefully acknowledged.