Rapid DNA barcoding-based fern and lycophyte inventories of protected areas—A pilot study to introduce a simple but effective protocol
DNA条形码技术实现保护区生物多样性的快速编目和鉴定——一种不过度依赖分类学家的快速高效的研究手段
Editor-in-Chief: Binbin Li
Handling Editor: Alison Nazareno
Abstract
enRecording inventories of species conserved in protected areas is a key step to evaluate the effectiveness of Kunming Montreal Global Biodiversity Framework (KM-GBF) targets, such as the expansion of protected areas. The application of DNA barcoding facilitates the rapid production of enables to obtain rapid inventories with reduced reliance on taxon experts. These inventories aim not only to confirm existing records but also to minimize gaps in our knowledge of the distribution and taxonomy of species targeted for conservation through the implementation of protected areas. This pilot study introduces a simplified DNA barcoding pipeline as a reliable tool for recording fern and lycophyte species occurring in protected areas. The pipeline emphasizes limited and/or short training requirements, reducing the input required from taxon experts and maximizing shared benefits between conservationists and taxonomists. Despite using a single DNA barcoding region, 78% of the accessions were unambiguously identified to the species level. This applied approach not only confirmed previous records but also identified several previously overlooked species, either as newly recorded species conserved in the protected area or as species new to science. The pilot project effectively documented known species diversity and identified gaps in our taxonomic knowledge by discovering previously unknown and locally rare taxa. This rapid assessment enhances productive exchanges between conservation practitioners and taxon experts, with substantial benefits for both parties.
摘要
zh了解和认识保护区内的生物多样性是开展科学研究和保护的前提,也是实现昆明-蒙特利尔公约目标的基础,但是分类学家人数和规模的不足是当前开展保护区生物多样性编目和分类研究的制约因素之一,而相对标准化的DNA条形码技术是一种不过度依赖分类学家的快速高效的研究手段。在保护区内开展深入的野外调查,并结合DNA条形码研究,不仅可以记录保护区内的常见物种,而且有助于发现珍稀物种或地区特有种。
本研究通过在西双版纳国家级自然保护区勐仑子片区的绿石林开展石松类和蕨类野外调查,并结合室内DNA条形码研究,对该保护区内的蕨类植物多样性进行编目和鉴定。我们的研究发现,本研究手段不仅能发现和记录大部分的常见物种,还记录到勐仑特有种1个、勐仑新纪录种1个和可能的新种1个。本研究的优势是,通过相对易培训和操作的DNA条形码技术,在不过度依赖类群专家的情况下,可以快速完成保护区的物种编目和鉴定,进而能够提升对保护区生物多样性的认知和物种保护。
本研究的结果表明,仅利用单个通用的DNA条形码片段(叶绿体rbcL片段)即可以实现对78%的物种的准确鉴定,通过结合标本检视和文献查阅,还记录到勐仑特有种1个、勐仑新纪录种1个和可能的新种1个。因此,本研究呼吁未来的保护区物种调查和生物多样性编目可以引入DNA条形码技术,以实现对保护区的生物多样性的认识和研究。【翻译:刘红梅】
Plain language summary
enRecording the biodiversity conserved in protected areas is a crucial cornerstone to assess their effectiveness. Unfortunately, assessing the diversity within protected areas has been a major challenge due to the scarcity of taxonomists who can provide reliable identification. Here, we introduce a simple DNA barcoding approach that can be established and carried out by conservation practitioners with minimal involvement of taxonomic experts. Notably, the records obtained through this approach can confirm the occurrence of previously recorded species, identify new species, and even detect gaps in our taxonomic knowledge. These multiple benefits were demonstrated through a pilot project that recorded the fern species diversity in a small conservation area in southwest China, utilizing a simplified DNA barcoding approach to minimize dependence on taxonomic experts.
通俗语言摘要
zh了解保护区内的生物多样性是开展研究和保护的前提,但是分类学家人数和规模的不足是当前开展保护区生物多样性编目和分类研究的制约因素之一。传统的生物多样性数据收集往往基于科研人员进行的科学考察,并主要依赖分类学家进行物种鉴定。在当前生物多样性丧失日益加剧的情况下,如何高效地开展生物类群的记录和研究成为日益紧迫的任务。DNA条形码技术是利用生物体DNA中一段保守片段对物种进行快速准确鉴定的技术,具有易操作、易标准化和相对客观的优势,分类学家可以用DNA条形码技术鉴定物种及物种间亲缘关系,修改已有的分类学结论。通过对年轻的非专业人员进行短期培训,开展DNA条形码研究,结合分类学家的最终把关和质量控制,可以实现对研究区域内生物多样性的快速编目和鉴定。因此,分类学家+DNA条形码技术的结合可以有效提升我们对地区生物多样性的认识,我们呼吁应在更大范围内推广和应用。
Practitioner points
en
-
This highly simplified DNA barcoding protocol can be easily applied to address the species identification bottleneck.
-
Communicating results with experts is crucial for confirming new species records and possibly identifying species that may be new to science.
-
DNA barcoding improves the documentation of known species while also expanding the number of rare species known to occur in protected areas.
实践者要点
zh
-
整合分类学家的专业知识和DNA条形码技术,能提高地区生物多样性编目的效率;
-
现代研究手段与科研人员专业知识的有机结合,有利于更全面地了解和认识生物多样性;
-
DNA条形码技术不仅能够记录地区常见种,也有助于发现潜在的新物种。
1 INTRODUCTION
Protected areas, such as national parks and forest reserves, are the cornerstone of global efforts to conserve species threatened by anthropogenic transformations of their environments including deforestation, urbanization, and global climate warming (Pacifici et al., 2020; Williams et al., 2022). As a consequence, expansion of protected areas has been set as one of the targets of the Kunming-Montreal Global Biodiversity Framework (Hughes, 2023; Watson et al., 2023). Evaluation of the effectiveness of already established or intended protected areas requires reliable inventories of the species conserved in the area protected (Rodrigues & Cazalis, 2020). However, current inventories often focus only on a few iconic organisms, such as large mammals, birds, and selected tree genera, leaving significant gaps in the data for the vast number of taxa contributing to the ecosystems (Rodrigues & Cazalis, 2020). Consequently, there is insufficient evidence to support the assumption that protected areas effectively slow the decline of species diversity across all branches of the tree-of-life (Justin Nowakowski et al., 2023). Besides the lack of first records, another major challenge is the absence of repeated occurrence confirmations necessary to enable the evaluation of the effectiveness of applied management strategies. To overcome these challenges, effective management of protected areas thus requires to set targets that aim to achieve periodic documentation of all or at least a broad range of organisms occurring in these areas. Unfortunately, this task is complicated by mapping the biodiversity, especially repeated recording has to tackle the scarcity of taxonomic expertise for many lineages of organisms (Engel et al., 2021; Wheeler et al., 2012). Given this limited availability of taxon experts for most lineages of the tree-of-life, assessments of the biodiversity in protected areas require the employment of methodologies that minimize the need for expert input while avoiding compromising the essential quality of species identification. Here, we addressed specifically the utilization of DNA barcoding that presents a viable solution to these challenges by facilitating rapid species inventories with minimal taxonomic expertise. Since its introduction, DNA barcoding has proven valuable in conservation biology despite certain challenges require to be taken into account (e.g., Liu et al., 2014; Pereira et al., 2021; Song et al., 2023). This study reports on a pilot project designed to establish a DNA barcoding workflow with specific objectives: (1) generating DNA sequences with limited costs and technological requirements, (2) simultaneously assessing and creating a reference data set, and (3) establishing an effective feedback loop between conservation practitioners and taxon experts.
DNA barcoding was introduced to leverage advancements in DNA sequencing technologies for achieve reliable species identification without the constant need for taxon experts involvement at every step (Hebert et al., 2003). Since its introduction, substantial efforts were not only taken to establish barcoding for all organisms, including land plants (e.g., CBOL Plant Working Group, 2009; Kress, 2017), and has evolved to incorporate to expand from the CBOL concept towards more flexible frameworks by integrating new DNA technologies such as metabarcoding (Ruppert et al., 2019). The successes and challenges of DNA barcoding for plants can be illustrated by its application to identify ferns. It has been utilized in a wide range of applications such as to improve quality control of fern fragments in the medicinal plant trade (Ma et al., 2010), to tackle the identification of fern gametophytes (Li et al., 2009; Nitta et al., 2017; Nitta & Chambers, 2021; Schneider & Schuettpelz, 2006), to document genetic differentiation among closely related fern species (Liu et al., 2018; Wang et al., 2016), and to enhance documentation of fern diversity in various habitats, from temperate (de Groot et al., 2011) to tropical regions (Ebihara et al., 2010; Nitta et al., 2020; Trujillo-Argueta et al., 2021). These studies had in common the requirement to utilize reference data sets established under the guidance of taxon experts. Some well-curated DNA barcode libraries are available, such as for woody plants in tropical and subtropical China (Jin et al., 2023) and flowering plants of northwestern China's arid regions (Song et al., 2023). However, the broader application of DNA barcoding for monitoring plant diversity in protected areas requires simultaneous inventory assessments and the creation of DNA reference barcoding libraries.
Here, we describe a pipeline to apply DNA barcoding to record the species diversity in protected areas, specifically focusing on the fern and lycophyte diversity of the Green Stone Forest Fragment that belongs to Menglun Sub-Nature Reserve of the Xishuangbanna National Forest Reserve, located in southwest China. Some information about the fern diversity of this fragment has been assessed by employing historical records—available in the form of herbarium specimens—in a previous study on the fern and lycophyte diversity of the Xishuangbanna Dai Autonomous Prefecture (Chen et al., 2022). Ferns and lycophytes are not only common in tropical forest habitats but are also considered excellent ecological indicators for evaluating such ecosystems (Della, 2022). The surveyed protected area is known to host local endemics, such as Cyrtomium latifalcatum S. K. Wu & Mitsuta (Wu & Mitsuta, 1985; Zhang et al., 2013). The pilot project is set up with the specific aim to be applicable without substantial training efforts and with access to laboratories providing only basic facilities for DNA extraction and DNA barcode amplification. Therefore, the number of DNA regions employed was restricted to a single region, the chloroplast genome-based rbcL gene. This gene has been the work-horse of fern and lycophyte phylogenetics since the earliest studies applied molecular tools to improve our understanding of their phylogenetic relationships (Nitta et al., 2022). As a consequence, more than 20,000 rbcL sequences of ferns and lycophytes have been deposited in public databases such as GenBank (https://www.ncbi.nlm.nih.gov/). To demonstrate the simplicity of the core parts of the pipeline, collecting surveys and molecular lab work were carried out by an intern student from a local university. The input from experts with extensive taxonomic knowledge was restricted to several essential steps, such as assembling a reference library and quality control. A specific target of the pipeline has been the establishment of feedback mechanisms to address taxonomic challenges.
Specifically, the results of the study were evaluated to satisfy the following requirements: (1) most species previously recorded for the protected area were recovered through unambiguous DNA identification; (2) species gaps in the reference library were successfully addressed by obtaining new sequences, especially of local endemics; and (3) short-comings of current species treatments were successfully detected, including the discovery of accessions that may represent species new to science.
2 MATERIALS AND METHODS
2.1 Study site
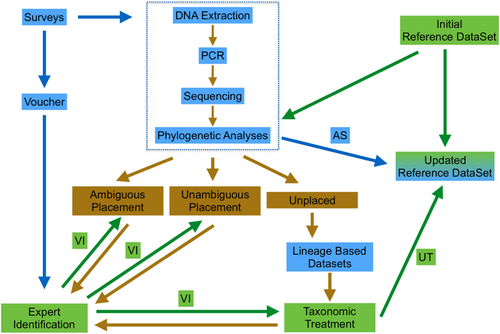
The pilot project employed a workflow designed to enable contributors with limited training in plant taxonomy and molecular biology to perform most of the data production, thereby reducing the dependence on experienced taxonomists and optimizing feedback between conservation practitioners and taxonomists (see Figure 1). Transect surveys of the Green Stone Forest Fragment (21.91022409° N, 101.28119972° E) within the Menglun Sub-Nature Reserve of the Xishuangbanna National Forest Reserve, located in the southern part of the Xishuangbanna Dai Autonomous Prefecture (Yunnan, China), were carried out repeatedly by students with minimal knowledge of the target plant lineage, without the involvement of taxon experts. The role of the experts was minimized to providing initial guidance for conducting surveys, training in the molecular laboratory, and identifying the accessions using traditional morphology-based approaches. All ferns and lycophytes were collected to (1) obtain voucher specimens and (2) obtain materials for DNA extraction. In addition, all collected samples were imaged in the field. Voucher specimens were generated using standard protocols and deposited in the research laboratory at the Xishuangbanna Tropical Botanical Garden, CAS. Pinnae or lamina fragments were removed, dried in silica gel, and stored in the laboratory. The remaining material was preserved for future studies. All images taken were transferred to an image database. Future surveys will only require the collection of leaf fragments, minimizing the impact on rare species by avoiding the collection of whole plants.
2.2 DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing
Whole genomic DNA was extracted using commercially available DNA extraction kits (Tiangen Biotech Co.) following the manufacturer's protocol (see File S1 for further details). The obtained DNA was then used to set up PCR reactions to amplify a single DNA fragment, a process commonly employed in DNA barcoding and molecular phylogenetics of these plants, specifically the chloroplast genome region rbcL (see File S1). The PCR products were sent to Sangon Biotech Co., Ltd for Sanger sequencing. The obtained sequences were assembled and stored using widely available and freely accessible software tools, such as BioEdit 7.1 (https://thalljiscience.github.io; Hall, 1999). All sequences were integrated into a single database in the commonly used nexus format. The pipeline of DNA extraction, PCR amplification, and DNA sequencing was summarized in File S1. All newly generated sequences were then checked for taxon identity using DNA Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to obtain a list of accessions with highly similar DNA sequences (see BLASTN query procedure; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). The reported list comprised information about the scientific name (genus, species), GenBank accession number, and several statistical values, particularly measures of sequence similarity, such as the percentage identity (Per. Ident) that enabled a ranking of the accessions with similarities of up to 100%. Any potential contamination and mismatch sequences were identified and filtered out by this first quality control step.
2.3 DNA barcodes reference library
A reference database was assembled for the same DNA region. For each species, all available rbcL sequences were downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/), which is freely accessible to the public. To avoid generating excessively large reference databases, up to three sequences per species were maintained, emphasizing accessions with recorded voucher information and relatively long reads (>1200 bp). These three accessions were filtered from all accessions available in early 2024 using the following criteria. Sequences generated early days of Sanger sequencing, thus before the year 2000, were avoided if possible. Specific attention was given to ensuring the exclusion of misidentified accessions. All sequences of a single region were assembled into a single nexus matrix and aligned using widely used and freely available alignment tools, with manual adjustment as necessary to handle poorly edited sequences. This part of the process was carried out using Mesquite 3.81 (https://www.mesquiteproject.org). Instead of focusing only on species recorded from the Menglun Sub-Nature Reserve of the Xishuangbanna National Forest Reserve, the assembled data set included all fern and lycophyte species previously recorded in the Xishuangbanna Dai Autonomous Prefecture (Chen et al., 2022), with some alterations reflecting recent taxonomic progress. The classification applied followed PPGI (2016) with minor alterations.
2.4 Sequence alignment and phylogenetic reconstruction
The matrix containing the newly generated sequences was merged with the reference matrix. The alignment of all sequences of the combined matrix was checked and adjusted manually using Mesquite. This matrix was then used for phylogenetic analyses employing PhyML (http://www.atgc-montpellier.fr/phyml/; Guindon et al., 2010). The optimal maximum likelihood hypothesis was recovered by applying automatic model selection via SMS, with the Bayesian Information Criterion selected (Lefort et al., 2017), and a starting tree obtained with BioNJ (Gascuel, 1997). Standard bootstrap analysis was conducted with up to 1000 replicates. The generated phylogenetic hypotheses were visually inspected using freely available tools designed to visualize phylogenetic trees, namely, Figtree 1.4.4 (http://tree.bio.ed.ac.uk/software/Figtree/) and ITOL v6 (https://itol.embl.de/itol.cgi; Letunic & Bork, 2021). The results were interpreted using the following protocol (see Figure 1). Accessions nested in clades formed by reference sequences identified as a single species were considered unambiguously identified. Clades formed by accessions representing more than one species were considered ambiguous. Accessions forming lineages not including any reference sequence were considered unknown. These accessions were assumed to represent either nonsampled species or species potentially new to science. All voucher specimens were identified by an experienced professional. These identifications were then compared to those obtained through the DNA barcoding process. The following categories were considered: (1) Identical identification, (2) conflicting identification, (3) species ambiguous and unplaced in the DNA barcoding but identified using morphology, and (4) species unplaced in the DNA barcoding and also unknown to science as of February 2024. To clarify the taxonomic status of accessions belonging to Category 4, all rbcL sequences of close relatives available in GenBank (February 2024) were downloaded and assembled into a matrix as described above. The phylogenetic analyses were run as previously described. This procedure was specifically carried out for the genera Cyrtomium J. Sm., Hymenasplenium Hayata, Leptochilus Kaulf., and Asplenium L., which are associated with several closely related species complexes occurring in China and adjacent regions.
3 RESULTS
The reference data set included 382 out of the 434 fern and lycophyte species recorded to occur in the Xishuangbanna Dai Autonomous Prefecture (see Table S1), resulting in a taxon coverage of 88.0%. Taxa without any published DNA sequences in public databases are primarily local endemics in Xishuangbanna, namely, Bolbitis confertifolia Ching, C. latifalcatum S. K. Wu & Mitsuta, Polystichum paradeltodon L. L. Ziang, Pteris menglaensis Ching, and Pteris undulatipinna Ching besides. Additionally, some Yunnan endemics such as Crepidomanes chui Ching & P. S. Chiu, Diplazium quadrangulatum (W. M. Chu). Z. R. He, and Lomagramma yunnanesis Ching. The reference data set did include some Xishuangbanna endemics, such as Arachniodes pseudoasssamica Ching and Leptochilus mengsongensis M. X. Zhao. Considering historical records specific to the Menglun Sub-Reserve, the reference data set covered 197 out of 183 species, achieving a coverage of 92.3%.
In total, 70 accessions were sampled during surveys and incorporated into the DNA barcoding test sample (Table 1). Employing the phylogenetic hypotheses generated (Figure 2), 55 accessions (78.6%) were unambiguously identified, whereas nine accessions (12.9%) were ambiguously identified (see Table 1). Finally, five accessions (7.1%) were unplaced, and one accession (1.4%) carried an incorrect accession number. In total, the survey recovered 53 species belonging to 26 genera, 14 families, and five orders (Table 1). Ambiguous identified accessions included Angiopteris helferiana C. Presl (accession CYR35, CYR41, CYR45), Adiantum ⨯ meishanianum F. S. Hsu ex Yea C. Liu & W. L. Chiou (accession CYR24), Asplenium humbertii Tardieu (accession CYR23), Christella jinghongensis (Ching) A. R. S. M. & S. E. Fawc. (accession CYR40), Leptochilus flexilobus (Christ) Liang Zhang & Li Bing Zhang (accession CYR57), Lygodium flexuosum (L.) Sw. (accession CYR72), and Pteris venusta Kunze (accession CYR04). The unplaced species included C. latifalcatum (accession CYR21), a local endemic that was missing in the reference data set. Four accessions were unplaced because they formed independent clades in the genera: Asplenium (accession CYR17), Hymenasplenium (accession CYR12, CYR59), and Leptochilus (accession CYR2) (see Figure 2).
| Lineage | Species | ID DNA BAR CAT | Acc. nr. | Record status |
|---|---|---|---|---|
| 1.1 | Selaginella bodinieri Hieron. | Unambiguous | CYR26 | New record |
| 1.1 | Selaginella helferi Warb. | Unambiguous | CYR34 | Confirmed |
| 1.1 | Selaginella picta A. Br. ex Baker | Unambiguous | CYR51, CYR67 | Confirmed |
| 1.1 | Selaginella repanda (Desv. Ex Poir.) Spring | Unambiguous | CYr14 | Confirmed |
| 2.1 | Angiopteris helferiana C. Presl | Ambiguous (4) | CRR35, CYR41, CYR45 | Confirmed |
| 2.2 | Crepidomanes latealatum (Bosch) Copel. | Unambiguous | CYR05 | Confirmed |
| 2.3 | Lygodium circinnatum (N. L. Burm.) Sw. | Unambiguous | CYR32 | Confirmed |
| 2.3 | Lygodium flexuosum (L.) Sw. | Ambiguous (6) | CYR72 | Confirmed |
| 2.4 | Microlepia rhomboidea (Wall. Ex Kunze) Prantl | Unambiguous | CYR48 | Confirmed |
| 2.5 | Adiantum caudatum L. | Unambiguous | CYR13, CYR64 | Confirmed |
| 2.5 | Adiantum menglianense Y. Y. Qian | Unambiguous | CYR11 | New record |
| 2.5 | Adiantum × meishanianum F. S. Hsu ex Yea C. Liu & W. L. Chiou | Ambiguous (3) | CYR24 | New record |
| 2.5 | Antrophyum wallichianum M. G. Gilbert & X. C. Zhang | Unambiguous | CYR30 | New record |
| 2.5 | Pteris arisanensis Tagawa | Unambiguous | CYR10 | Confirmed |
| 2.5 | Pteris biaurita L. | Unambiguous | CYR52 | Confirmed |
| 2.5 | Pteris ensiformis N. L. Burm. | Unambiguous | CYR33 | Confirmed |
| 2.5 | Pteris venusta Kunze | Ambiguous (1) | CYR04 | Confirmed |
| 2.6 | Asplenium humbertii Tardieu | Ambiguous (2) | CYR23 | New record |
| 2.6 | Asplenium nidus L. | Unambiguous | CYR20 CYR63 | Confirmed |
| 2.6 | Asplenium saxicola Rosenst. | Unambiguous | CYR25 | Confirmed |
| 2.6 | Asplenium spec. | Unplaced | CYR17 | New spec |
| 2.6 | Hymenasplenium excisum (C. Presl) S. Lindsay | Unambiguous | CYR46 | Confirmed |
| 2.6 | Hymenasplenium spec. unknown | Unplaced | CYR12, CYR59 | New spec |
| 2.6 | Anisocampium cuspidatum (Bedd.) Yea C. Liu, W. L. Chiu & M. Kato | Unambiguous | CYR06 | Confirmed |
| 2.6 | Diplazium alatum (Christ) R. Wei & X. Z. Zhang | Unambiguous | CRY53 | Confirmed |
| 2.6 | Diplazium simile (W. M. Chu) R. Wei & X. C. Zhang | Unambiguous | CRY36, CYR50 | New record |
| 2.6 | Abacopteris nudata (Roxb.) S. E. Fawc. & S. R. Sm. | Unambiguous | CYR60 | Confirmed |
| 2.6 | Christella dentata (Forssk.) Brownsey & Jermy | Unambiguous | CYR38 | Confirmed |
| 2.6 | Christella jinghongensis (Ching ex K. H. Shing) A. R. Sm. & S. E. Fawc. | Ambiguous (5) | CYR40 | Confirmed |
| 2.6 | Christella parasitica (L.) H. Lév. | Unambiguous | CYR39 | Confirmed |
| 2.6 | Christella subelata (Baker) Holttum | Unambiguous | CYR37 | Confirmed |
| 2.6 | Reholttumia truncata (Poir.) S. E. Fwac. & A. R. Sm. | Unambiguous | CYR47 | Confirmed |
| 2.7 | Davallia griffithiana Hook. | Unambiguous | CYR31 | Confirmed |
| 2.7 | Bolbitis scandens W. M. Chu | Unambiguous | CYR42, CYR70 | New record |
| 2.7 | Cyrtomium latifalcatum S. K. Wu & Mitsuta | Unplaced | CYR21 | Confirmed |
| 2.7 | Nephrolepis falciformis J. Sm. | Unambiguous | CYR08 | Confirmed |
| 2.7 | Drynaria bonii Christ | Unambiguous | CYR03, CYR29 | New record |
| 2.7 | Lepisorus carnosus (J. Sm.) C. F. Zhao, R. W. & X. C. Zhang | Unambiguous | CRY15 | Confirmed |
| 2.7 | Lepisorus zippelii (Blume) C. F. Zhao, R. Wei, & X. C. Zhang | Unambiguous | CYR65, CYR68 | Confirmed |
| 2.7 | Leptochilus flexilobus (Christ) Liang Zhang & Li Bing Zhang | Ambiguous (6) | CYR56, CYR57 | New record |
| 2.7 | Leptochilus spec. unknown | Unplaced | CYR02 | New spec |
| 2.7 | Microsorum cuspidatum (D. Don) Tagawa | Unambiguous | CYR28 | Confirmed |
| 2.7 | Microsorum punctatum (L.) Copel. | Unambiguous | CYR27 | Confirmed |
| 2.7 | Pyrrosia nuda (Giesenh.) Ching | Unambiguous | CYR22 | Confirmed |
| 2.7 | Pyrrosia nummulariifolia (Sw.) Ching | Unambiguous | CYR16 | Confirmed |
| 2.7 | Pteridrys cnemidaria (Christ.) C. Chr. & Ching | Unambiguous | CYR09, CYR49, CYR55 | Confirmed |
| 2.7 | Tectaria devexa (Kunze) Copel. | Unambiguous | CYR19 | Confirmed |
| 2.7 | Tectaria fauriei Tagawa | Unambiguous | CYR43 | Confirmed |
| 2.7 | Tectaria fuscipes (Wall. ex Bedd.) C. Chr. | Unambiguous | CYR44, CYR62, CYR66 | Confirmed |
| 2.7 | Tectaria herpetocaulos Holttum | Unambiguous | CYR07, CYR58 | Confirmed |
| 2.7 | Tectaria impressa (Fee) Holttum | Unambiguous | CYR18, CYR71 | Confirmed |
| 2.7 | Tectaria quinquefida (Baker) Ching | Unambiguous | CYR61 | Confirmed |
| 2.7 | Tectaria zeylanica (Houtt.) Sledge | Unambiguous | CYR01 | Confirmed |
- Note: Columns report: lineage (linear clade number); species according to expert identification and DNA barcoding identification; ID DNA BAR CAT = Identification of DNA barcoding category as unambiguous, ambiguous, unplaced, confused sample (numbers see below); Acc. Nr. = Accession Number (number of the accessions collected in surveys of the Green Stone Forest Fragment); Record Status = species presented records for the Menglun Sub-Nature Reserve of which the Green Stone Forest Fragment is part of as previously recorded (confirmed), recorded for the first time (new record), or a perhaps a species new to science (New spec). Causes of ambiguous DNA barcoding identifications: (1) lack of differentiation among Pteris heteromorpha, Pteris subquinata, and Pteris venusta; (2) lack of differentiation between Asplenium antrophyoides and Asplenium humbertii; (3) shared chloroplast DNA between Adiantum × meishanianum and its maternal parent Adiantum malesianum; (4) lack of differentiation among species of Angiopteris; (5) lack of differentiate between Christella jaculosa and Christella jinghongensis in the two available rbcL sequences; and (6) lack of differentiation among Lygodium flexuosum and Lycodium salicifolium in rbcL.
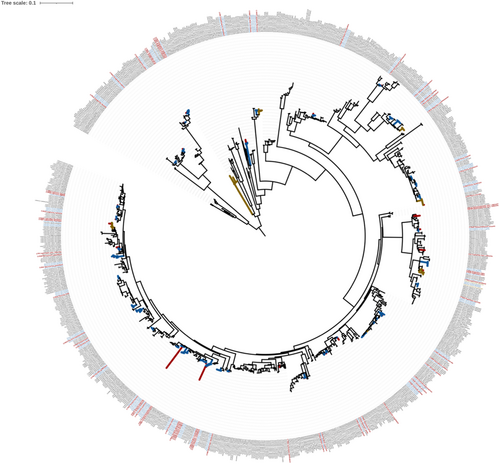
4 DISCUSSION
In total, the study analyzed 70 accessions using the DNA barcoding protocol, resulting in the identification of 41 species previously recorded for the Menglun Sub-Nature Reserve, and nine species that were newly recorded. Among the newly recorded species, Asplenium humbertii Tardieu was documented for the first time in the Xishuangbanna Dai Autonomous Prefecture. Additionally, four accessions required further study due to unresolved species identities. The DNA barcodes determined their generic relationship but did not match them with previously described species, suggesting the presence of three species potentially new to science (see Table 1, New spec). Two of these accessions were identified as a sister pair within the genus Hymenasplenium, while the other two were placed in the genera Asplenium and Leptochilus. Furthermore, the study provided the first DNA sequences for the local endemic C. latifalcatum.
The pilot project demonstrated the effectiveness of a straightforward DNA barcoding approach, utilizing a single plastid-based DNA fragment, such as rbcL, to reliably identify approximately 75% of the accession recovered. The success rates of DNA barcoding in this study were comparable to previous reports on the DNA barcoding of ferns and lycophytes in Japan (Ebihara et al., 2010) and French Polynesia (Nitta et al., 2017). Unfortunately, some common challenges of DNA barcoding were encountered, including unplaced species and ambiguous identifications. Ambiguous identification is a recurring issue due to DNA barcodes often failing to differentiate species complexes as a consequence of hybridization, polyploid speciation, and slow mutation rates. For instance, the latter process has been reported for the marattioid genus Angiopteris Hoffm. (Lehtonen et al., 2020; May et al., 2021). Our results are consistent with the expectation that low mutation rates result in failed species identification when utilizing only rbcL sequences. Hybridization led to the ambiguous identification of accession CYR24 as Adiantum × meishanianum F. S. Hsu ex Yea C. Liu & W. L. Chiou (Shang et al., 2016). DNA barcodes utilizing genes from the uniparentally inherited chloroplast genome will fail to provide unambiguous identification of taxa arising from hybridization or polyploidization (Liu et al., 2018). The other ambiguously identified accessions are currently unplaced and require further study. For example, three out of the four accessions belonging to the genus Christella H. Lev. were identified unambiguously as Christella dentata (Forssk.) Brownsey & Jermy, Christella parasitica (L.) H. Lev., and Christella subelata (Baker) Holttum, whereas the fourth accession was ambiguously identified according to the rbcL sequences as Christella jaculosa (Christ) Holttum or Christella jinghongensis (Ching ex K. H. Shing) A. R. Sm. & S. E. Fawc. Both species are known to occur in Xishuangbanna (Li et al., 2013) and were represented by a single rbcL sequence in the reference data set. Considering morphological diagnostic characteristics (Li et al., 2013), the accession was identified as C. jinghongensis. Despite significant progress in the morphological diagnostics of this genus (Li et al., 2013), the species identity of specimens utilized to generate DNA sequences requires verification. Attention to taxonomic ambiguities is especially crucial in cases where only one rbcL sequence is available. Accessions belonging to Lepisorus sect. Lemmaphyllym (C. Presl) C. F. Zhao, R. Wei & X. C. Zhang (Zhao et al., 2020) exemplify the need for further taxonomic clarification in southern Yunnan (Wei & Zhang, 2013; Zhao et al., 2020). Accessions identified as Leptochilus flexilobus illustrate the need for continued study of the confusing taxonomy of these ferns (Zhang et al., 2019).
4.1 New records
During the survey, accessions were encountered that were identified as A. humbertii (see Lin & Viane, 2013). This species has previously been recorded from limestone rocks in southeastern Yunnan, as well as in Guangxi, Hainan Island, Laos, Thailand, and Vietnam. It is considered to form a species complex with Asplenium antrophyoides Christ and Asplenium grevillei Wall. ex Hook. & Grev., which requires further study (Dong, 2011; Lin & Viane, 2013; Wei & Dong, 2012). Cytological and DNA sequence data are available for only two of these three species: the tetraploid A. antrophyoides and the hexaploid A. humbertii (Dong, 2011; Schneider et al., 2017; Xu et al., 2020). The sample from the Green Stone Forest Fragment was nested in a clade comprising accessions of both species but failed to resolve the differentiation between the two species. Therefore, further studies are required to clarify the status of these accessions. Here, we accept the identification based on the treatment of these ferns in the Flora of China (Lin & Viane, 2013). As a consequence, the range of A. humbertii now includes not only southeastern Yunnan but also southwestern Yunnan.
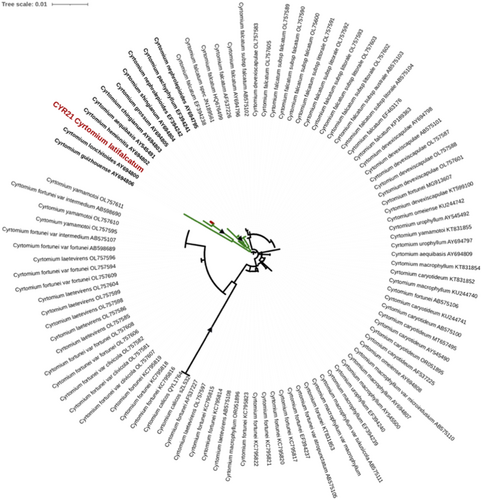
4.2 New genetic resources of the local endemic C. latifalcatum
C. latifalcatum is currently known only from Xishuangbanna, Yunnan (Wu & Mitsuta, 1985; Zhang et al., 2013). Previous records support occurrences of this local endemic in the Mengla Sub-Nature Reserve (Chen et al., 2022). The generated phylogenetic hypothesis recovered this species as part of a clade comprising species known to occur in southern China (Figure 3). Several of these species are local endemics, such as the tetraploid Cyrtomium chingianum P. S. Wang and the diploid Cyrtomium guizhouensis H. S. Kung & P. S. Wang. All species of this clade are known to occur in limestone rocks (Zhang et al., 2013). The clade also shows notable variation in ploidy levels, including diploids, triploids, and tetraploids, as well as sexual and apomictic reproduction (Lu et al., 2006; Zhang et al., 2013). Similar to other fern genera occurring on karst formations in southern China and northern Vietnam, several species new to science have been discovered in recent years, such as Cyrtomium calcis Liang Zhang, N. T. Liu & Li Bing Zhang (Lu et al., 2023) and Cyrtomium remotipinnum Yan Liu & H. J. Wei (Nong et al., 2023). The majority of the species related to C. latifalcatum share narrow distribution ranges due to their ecological specialization to karst formations. Therefore, these species are vulnerable to extinction threats caused by anthropogenic activities and require special attention. C. latifalcatum is considered to be endangered (Chen et al., 2022).
4.3 Unplaced accessions
Accessions recovered as unplaced species require further exploration because they highlight recording gaps that are not restricted to the lack of documentation of already known species but represent species putatively new to science. Each of these cases will be discussed more extensively, although we are not attempting to resolve the taxonomic issues here.
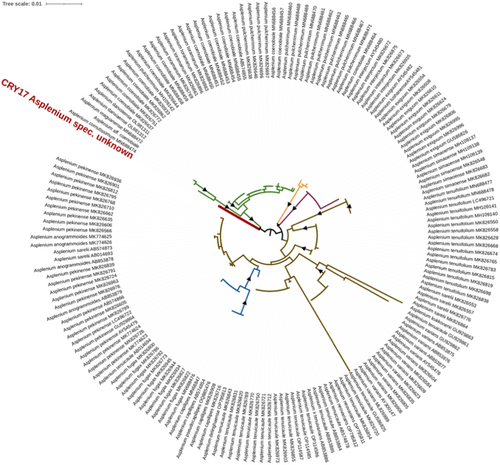
One accession (accession CYR17) was found to form an independent clade sister to the Asplenium coenobiale complex (Figure 4). This complex comprises currently five known species (Jiang et al., 2011; Siqi et al., 2019; Xu et al., 2022). Two widespread species, namely, A. coenobiale Hence and Asplenium pulcherrimum (Baker) Ching, have been recognized since the exploration of Southeast Asian fern diversity about 100 years ago. In the last 20 years, only three local endemics have been reported: Asplenium cornutissimum X. C. Zhang & R. H. Jiang from Guangxi in 2011, Asplenium maguanense S. Q. Liang, R. Wei & X. C. Zhang from southeastern Yunnan in 2019, and Asplenium danxiaense K. W. XU from Guangdong in 2022. These new discoveries arguably indicate the need for a comprehensive investigation into the diversity of these ferns in southern China and adjacent tropical regions, focusing specifically on Karst and Danxia outcrops, as these species have been reported from such habitats (Fu et al., 2022; Jiang et al., 2011; Siqi et al., 2019; Xu et al., 2022). The taxonomic status of accession CYR17 is currently under scrutiny using additional samples obtained in the Menglun region.
Other notable discoveries from our survey were the two accessions (CYR12, CYR59) nested in Hymenasplenium (Figure 5). The taxonomy of the Old World representatives of this genus has undergone substantial revisions in recent years. In particular, the concept of Hymenasplenium unilaterale as a Paleotropical species has been dismissed. Previously rejected species have been reinstated, and several new species have been described (e.g., Chang et al., 2018, 2022; Lin & Viane, 2013; Xu et al., 2018; Zhang et al., 2021). In addition to the taxonomic revisions within the H. unilaterale complex, eight species of this genus have been documented to occur in the Xishuangbanna Dai Autonomous Prefecture: Hymenasplenium apoganum (N. Kurak. & Hatan.) Nakaike, Hymenasplenium cheilosorum (Kunze ex Mett.) Tagawa, Hymenasplenium excisum (C. Presl) S. Linds., H. laterepens N. Murak. & Cheng ex Y. Fen Chang & K. Hori, Hymenasplenium obscurum (Blume) Tagawa, Hymenasplenium obtusidentatum Y. Fen Chang & G. Cheng Zhang, and Hymenasplenium pseudobscorum Viane. Notably, two of these species were introduced with type specimens collected from this area, namely, H. laterepens (Chang et al., 2018) and H. obtusidentatum (Chang et al., 2022). The two accessions recovered as an independent clade are morphologically distinct from all these species but resemble somewhat collections reported in a cytological study on Hymenasplenium in Xishuangbanna (Cheng & Murakami, 1998). These authors reported a diploid of Hymenasplenium as Hymenasplenium latipinnum, but this name was not properly published. Thus, Lin & Viane (2013) reported these samples as Hymenasplenium subnormale (Copel.) Nakaike, yet pointed out clear differences between the specimens from Yunnan and those from the type locality in the Philippines. These specimens require further comparative study to clarify their taxonomic status.
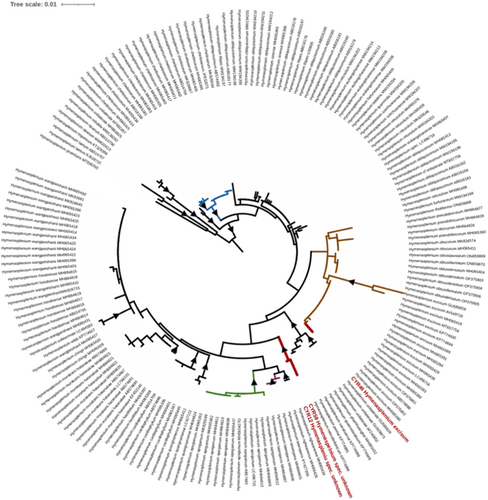
Recovering the accession CYR2 of Leptochilus not fitting to any published species (Figure 6) aligns with recent progress in the taxonomic studies of this primarily southeast Asian genus (Wei et al., 2023; Yu et al., 2024; Zhang et al., 2019; Zhao et al., 2017). These studies not only introduced several new species but also questioned previous taxonomic treatments of the genus. The accession CYR2 clustered in a clade (Figure 6) containing accessions of an unknown species, namely, Zhang et al. 6377 (GenBank accession number MH768422), Zhang et al. 7365 (GenBank accession number MH768436), and Zhang et al. 6711 (GenBank accession number MH768459). These three collections were obtained in central and northern Vietnam (Zhang et al., 2019). Therefore, our accessions extend the range of this clade from Vietnam to southwest Yunnan.
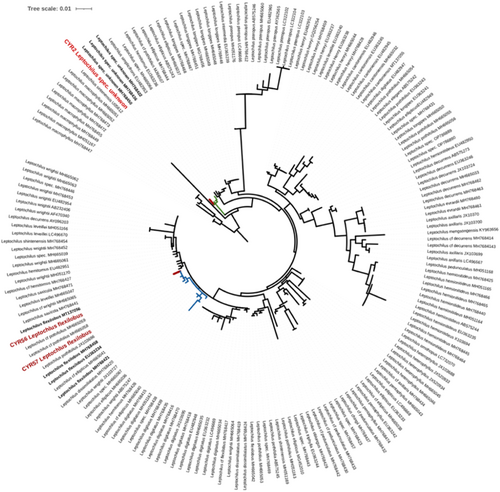
4.4 DNA barcoding-based inventories
As stated in the introduction, this study was designed as a pilot to explore the benefits and challenges of utilizing DNA barcoding to record the species diversity of protected areas with minimal involvement of taxonomists with expertise in fern taxonomy. Surprisingly, the surveys carried out by inexperienced students recorded common species as expected but also identified rare taxa. In particular, the recovery of C. latifalcatum is of importance. Repeated surveys documented a total of six individuals of this local endemic in the Green Stone Forest Fragment, which is its type locality (Wu & Mitsuta, 1985). Recovering this species confirmed its continued presence in its type locality and will also facilitate the establishment of effective conservation measurements for its protection. Given the small number of individuals recovered, there is a need to develop a comprehensive program to protect this species. Such a program should include recording crucial evidence required including assessments of ecological preferences, assessing demography, and genetic diversity, and collecting spores for ex situ reproduction followed by reintroduction (Wall et al., 2024).
An important lesson from this study was the confirmation of the concept's effectiveness. Non-Taxon expert collection was found to be effective, although feedback loops with taxonomists are essential to take full advantage of the evidence obtained. The recovery of new records and species new to science also highlighted the significant gaps in our records of species occurring in protected areas. In the case of the Green Stone Forest Fragment, this result may appear surprising given the history of plant diversity exploration in southern Yunnan (see Chen et al., 2022) and the proximity of this fragment to the core research center of the Xishuangbanna Tropical Botanical Garden. However, it is consistent with the ongoing discovery of new species in this region, such as Hymenasplenium laterepens N. Murak. & Cheng ex Y. Fen Chang & K. Hori (Chang et al., 2018), Leptochilus mengsongensis M. X. Zhao (Zhao et al., 2017) and Polystichum menglaense Z. L. Liang & Li Bing Zhang (Liang et al., 2021). Furthermore, recent studies have provided evidence supporting the assumption that these diversity-rich tropical forests are home to a disproportional frequency of rare fern and lycophyte species (Cicuzza, 2021). The approach introduced here has the potential to overcome hurdles that have hampered efforts to assemble comprehensive inventories of protected areas, even for those that are easily accessible.
5 CONCLUSIONS
Our pilot study demonstrated the effectiveness of a simplified DNA barcoding approach to obtain rapid assessments of the fern and lycophyte diversity in protected areas. sampled accessions were successfully identified, although some ambiguous identifications and unplaced accessions still require expert intervention. A significant challenge is the limited availability of reliable identified rbcL sequences for many species. Only 33.4% of the species were represented with the three targeted reference sequences, while incomplete representation was achieved for 16.1% with two sequences, 38.5% with one sequence, and 12.0% of the species had no available DNA sequence. Thus, the simultaneous assembly of reference data alongside inventories is arguably the only realistic approach, but this procedure requires the involvement of taxon experts. Furthermore, the inventory illustrates the incompleteness of our current understanding of both the spatial distribution of species—evidenced by new records—and the taxonomy of plants occurring in species-rich areas like southern Yunnan. Lastly, the effectiveness of inventories can be further enhanced by linking them to simultaneously established image reference data sets, providing conservation practitioners with an additional toolset for species identification. The combination of DNA barcoding and digital imaging holds the promise of overcoming limitations specific to each approach, thereby ensuring the quality of the identifications obtained.
AUTHOR CONTRIBUTIONS
Hongmei Liu: Conceptualization; data curation; identification; formal analysis, methodology; investigation; project administration; writing—original draft; writing—review and editing. Yarong Chai: Data curation; investigation. Harald Schneider: Conceptualization; validation; identification; writing—original draft; writing—review and editing.
ACKNOWLEDGMENTS
The authors are grateful to the management of the Menglun subnature reserve of the Xishuangbanna National Forest Reserve for the permission to survey the Green Stone Forest Fragment. The authors acknowledge the financial support by the Yunnan Province Science and Technology Department (202101AS070012), Yunnan Revitalization Talent Support Program “Innovation Team” Project (202405AS350019), and 14th Five-Year Plan of the Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (E3ZKFF8B01). The authors would like to thank several team members who involved and helped in the field. The molecular work was supported by Institutional Center for Shared Technologies and Facilities of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (CAS).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
The research does not involve any experiments involving animals or humans.
Open Research
DATA AVAILABILITY STATEMENT
All newly generated DNA sequences were deposited in GenBank. The reference data set is available from the request to the corresponding author. All other accumulated data were summarized in the supporting information files.




