RETRACTED: Circ_0087199 depletion attenuates lipopolysaccharides-induced human periodontal ligament cell injury through the miR-527/TLR4 axis
Abstract
Background
Circular RNAs participate in the development of periodontitis. The present work aims to reveal the role and mechanism of circ_0087199 in human periodontal ligament cell (PDLC) injury during periodontitis.
Methods
PDLCs were treated with lipopolysaccharides (LPS) to establish a periodontitis cell model. Quantitative real-time polymerase chain reaction was used to detect the expression of circ_0087199, miR-527, toll-like receptor 4 (TLR4). Western blot analysis assay was performed to assess protein expression. Cell viability, proliferation, apoptosis and inflammation were investigated by cell counting kit-8, EdU assay, flow cytometry and enzyme-linked immunosorbent assay, respectively. Oxidative stress was evaluated by malondialdehyde assay kit and superoxide dismutase activity assay kit. The interaction between miR-527 and circ_0087199 or TLR4 was confirmed by a dual-luciferase reporter assay.
Results
Circ_0087199 and TLR4 expression levels were significantly increased, while miR-527 was decreased in the periodontal ligament tissues of periodontitis patients and LPS-stimulated PDLCs when compared with controls. LPS treatment inhibited cell viability and proliferation but induced cell apoptosis, inflammation and oxidative stress, whereas these effects were attenuated after circ_0087199 knockdown. Circ_0087199 bound to miR-527 and regulated LPS-caused PDLC damage by targeting miR-527. Additionally, the overexpression of TLR4, a target gene of miR-527, rescued miR-527 mimic-mediated effects on LPS-treated PDLCs. Further, the regulation of circ_0087199 toward TLR4 involved miR-527.
Conclusion
Circ_0087199 knockdown attenuated LPS-induced apoptosis, inflammation and oxidative stress of PDLCs by regulating the miR-527/TLR4 pathway.
1 INTRODUCTION
Periodontitis is a widespread inflammatory disease featured by progressive destruction of the periodontal ligaments and alveolar bone.1 This disease occurs as an immune response to dental plaque, and genetic and epigenetic factors also mediate its progression.2 In terms of aetiology, virulence factors such as lipopolysaccharide (LPS) stimulate macrophages and inflammatory cells to produce proinflammatory cytokines, leading to the production of matrix metalloproteinases and receptor activator of NF-κB ligand, finally destructing collagen fibers in periodontal tissues and causing osteoclast genesis and maturation.3 More than 10% of the world population (about 743 million individuals) are diagnosed with periodontitis.3 Moreover, periodontitis is associated with the occurrence of certain non-communicable chronic diseases.4 Additionally, the loss of periodontal support can reduce masticatory performance.5
Epigenetic factors, especially noncoding RNAs, are important players in periodontitis pathogenesis.6 Among the noncoding RNAs, circular RNAs (circRNAs) participate in progression of a variety of diseases by sponging microRNAs (miRNAs) or regulating alternative splicing.7 In addition, circRNAs are able to sequester proteins in specific locations.8 There is an emerging understanding of circRNA function in periodontitis pathogenesis. For example, circCDR1as expression was upregulated after osteogenic differentiation, and circCDR1as depletion reduced osteogenic gene expression and inhibited bone formation,9 confirming the involvement of circCDR1as in osteoblast differentiation in periodontal ligament stem cells. Circ_0062491 overexpression ameliorated the increased apoptosis and inflammation of human periodontal ligament cells (PDLCs) by LPS, and mechanism assays showed that these effects were mediated by the miR-498/suppressor of cytokine signaling 6 axis.10 Another study revealed that gingival tissues of periodontitis patients exhibited a high circ_0087199 expression,11 however, the role of this circRNA in periodontitis pathogenesis remains unknown.
MiRNAs are noncoding ribonucleic acid molecules relevant to different physiological and pathogenic events. MiRNAs function as posttranscriptional regulators and can cause translational inhibition and transcript degradation of target messenger RNA by the mean of base-pairing interactions.12 MiRNAs are emerging players in periodontitis and are phenotypically and functionally associated with periodontitis progression. Considerable evidence showed that miRNAs initiate odontogenic differentiation and promote inflammatory responses after periodontitis.13 In addition, certain bacterial components induce the change of miRNA expression, and dysregulated miRNAs regulate the production of inflammatory factors in the periodontal pocket, mediate the expression of intercellular cell adhesion molecule-1 and E-Selectin, negatively modulate the TLR signaling pathway and NF-κB pathway, and promote neo-angiogenesis in periodontitis.14
Online databases predicted that miR-527 contained the binding sites of circ_0087199 and toll-like receptor 4 (TLR4). Whether circ_0087199 participates in periodontitis progression by regulating miR-527 and TLR4 has not been reported. Thus, the present work assembled the circ_0087199/miR-527/TLR4 axis to reveal the molecular mechanism relevant to circ_0087199 in periodontitis. The study was investigated using LPS-induced periodontitis cell model. The circ_0087199/miR-527/TLR4 axis was identified by a dual-luciferase reporter assay. The roles among circ_0087199, miR-527 and TLR4 in regulating LPS-induced periodontitis cell injury was analyzed using quantitative real-time polymerase chain reaction (qRT-PCR), Western blot analysis analysis, cell counting kit-8 (CCK-8), 5-Ethynyl-2′-deoxyuridine (EdU) assay, flow cytometry, enzyme-linked immunosorbent assay (ELISA), malondialdehyde (MDA) assay kit and superoxide dismutase (SOD) activity assay kit. The work aimed to expand our current understanding of periodontitis occurrence and explore novel therapeutic avenues for periodontitis.
2 MATERIALS AND METHODS
2.1 Periodontal ligament tissues
All subjects provided informed consent. Periodontitis patients (N = 31) with no systemic diseases and age-matched volunteers (N = 31) with orthodontic requirements provided periodontal ligament tissues at 1st Medical Center of people's liberation army (PLA) General Hospital. These clinical samples were used for gene expression analysis and human PDLC isolation, and stored at −80°C. Sample collection was approved by the Ethics Committee of 1st Medical Center of PLA General Hospital.
2.2 PDLC isolation and LPS stimulation
Periodontal ligament tissues isolated from age-matched volunteers were minced into small pieces and incubated with collagenase (type I) (Life-iLAB biotech) in α-minimum essential medium (Sigma) under 37°C for 0.5 h. Enzymic digestion was performed to obtain single-cell suspensions, which were subsequently cultured in α-minimum essential medium supplemented with 10% fetal bovine serum (Sigma), 1% penicillin/streptomycin (Sigma) and 2 nM l-glutamine (Sigma) under 37°C with 5% CO2. PDLCs passed three to six were used for the present study. For functional experiments, the isolated cells were pretreated with 10 μg/mL LPS (Sigma) for 3 h to mimic the condition of periodontitis in vitro.10 Phosphate buffer solution (Sigma)-treated PDLCs were used as normal controls.
2.3 Cell transfection
Ribobio Co., Ltd. designed and provided the small interfering RNA of circ_0087199 (si-circ_0087199, target sequence 5′-GCTCCAAAAGAACTGAGTTCT-3′), miR-527 mimics (5′-CUGCAAAGGGAAGCCCUUUC-3′) and inhibitors (5′-GAAAGGGCUUCCCUUUGCAG-3′), and matched controls including si-NC, miR-NC, and in-miR-NC. TLR4 overexpression plasmid (TLR4, built using the 71-2590 bp of TLR4 gene) was built through enzyme digestion and ligation, and pcDNA 3.1(+) vector (EK-Bioscience) was used during the plasmid construction. PDLCs were maintained in 12-well plates before transfection and treated with transfection mixtures including EZ Trans Lipo reagent (Life-iLab biotech), DNA, miRNA mimics, miRNA inhibitors, and siRNAs for 20 min. Cell incubation was then performed under 37°C with 5% CO2.
2.4 qRT-PCR
Eighty milligram of periodontal ligament tissues and 5 × 106 PDLCs were treated with 1 mL of TRIzol (Life-iLab biotech) for RNA isolation. Five μg of RNA was treated with RNase R (4 U/μg RNA, Xiyuan Biotech) at 37°C for 0.5 h and then incubated at 65°C for 20 min to analyze circRNA stability. cDNA first chain synthesis reagents (Jiancheng biotech) were used for cDNA synthesis. Subsequently, primers (listed in Table 1), miRNA fluorescence quantitative detection reagents (Jiancheng Biotech), and 2 × qPCR Mix (Life-iLab biotech) were used for quantitation analysis. After normalization with internal references (U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with constant expression), gene expression was determined using the method.
| Name | Primer sequences for qRT-PCR (5′-3′) | |
|---|---|---|
| Circ_0087199 | Forward | TCAGAAATGCTCCAAAAGAACTGAG |
| Reverse | CACTCCAAGAGTGGCAGGTC | |
| TLR4 | Forward | AGACCTGTCCCTGAACCCTA |
| Reverse | CTCCCAGAACCAAACGATG | |
| miR-527 | Forward | ATGCATCTGCAAAGGGAAGC |
| Reverse | CTCAACTGGTGTCGTGGA | |
| U6 | Forward | CTCGCTTCGGCAGCACATA |
| Reverse | CGAATTTGCGTGTCATCCT | |
| GAPDH | Forward | CAAATTCCATGGCACCGTCA |
| Reverse | GACTCCACGACGTACTCAGC |
2.5 Nucleocytoplasmic separation assay
Nucleocytoplasmic separation of PDLCs was performed based on the standard procedures of Nuclear/Cytosol Fractionation Kit (BioVision). Briefly, PDLCs and CEB-A Mix were added into tubes, and the cell pellet was fully resuspended by vortex on the highest setting. Then, Cytosol Extraction Buffer-B was added into the tubes and centrifuged on the highest setting. The supernatant was immediately transferred to prechilled tubes, and the pellets were resuspended for circ_0087199 expression analysis.
2.6 Cell viability assay
Each 96-well plate was inoculated with PDLC suspension following LPS stimulation and transfections according to CCK-8 Kit (Life-iLab biotech). After a 48-h culture, CCK-8 reagent was added carefully, and the 96-well plates were incubated for 3 h under 37°C with 5% CO2. Enzyme immunoassay analyzer (Thermo Fishe) was used for sample analysis.
2.7 Cell proliferation analysis
EdU solution was diluted and heated in advance as per the guidebook of a cell proliferation imaging analysis kit (Life-iLab biotech). The treated EdU reagent was then added to each well for 2 h, and EdU residue that was not incorporated with DNA was removed by washing cells using phosphate buffer solution. The cells were subjected to fixation and penetration, followed by incubation with dye reaction solution and DAPI reagent (Sigma). Cell samples were analyzed using fluorescence microscope (Olympus).
2.8 Cell apoptosis assay
PDLCs treated as above were subjected to analysis based on the standard procedures of Annexin V-FITC Apoptosis Analysis Kit (Life-iLab biotech). In brief, PDLCs were resuspended in Annexin V-FITC binding solution before staining with Annexin V-FITC and propidium iodide. At last, flow cytometry was applied to quantify apoptotic cells.
2.9 ELISA
Three commercial human ELISA kits (#AC16088, #AC16102, and #AC16138, ACMEC Biotech) were used for IL-1β, IL-6 and TNF-α level analysis in cell culture supernatants according to the responding guidebooks. In brief, cell debris in cell culture supernatants was removed by centrifugation, and samples and standards were incubated as directed. After incubation with biotin-conjugate antibodies and streptavidin-HRP, test wells were read by an enzyme immunoassay analyzer (Thermo Fisher) within 30 min.
2.10 SOD activity assay
PDLCs treated as above were collected and broken with ultrasound in extraction solution. Subsequently, test wells were added with different reagents and/or samples as instructed in the manual of a SOD activity analysis kit (ACMEC Biotech). Spectrophotometer (Thermo Fisher) preheated for 30 min or more was used for sample analysis.
2.11 MDA assay
PDLCs treated as above were collected and broken with ultrasound in extraction solution as instructed in the manual of a MDA assay kit (ACMEC Biotech). MDA working solution, distilled water, and samples were added to test wells and then analyzed using a spectrophotometer (Thermo Fisher).
2.12 Dual-luciferase reporter assay
Wild-type and mutant reporter plasmids including circ_0087199 WT, TLR4 3′UTR WT, circ_0087199 WT, and TLR4 3′UTR WT were built using pmirGLO vector (Miaoling biotechnology) following the prediction of binding sites of miR-527 for circ_0087199 and TLR4 through circRNAs interactome and Targetscan online databases. These plasmids were transfected into PDLCs with miR-527 mimics or mimic control, and the culture medium was removed from the cells into which the reporter plasmids had been transferred. Lysis buffer diluted with sterilized water (Sigma) was used to lysate cells. After centrifugation, the supernatant was utilized for luciferase activity analysis as instructed in the manual of a commercial dual luciferase reporter assay kit (Life-iLab biotech).
2.13 Western blot analysis analysis
Periodontal ligament tissues and PDLCs were mixed with RIPA buffer (Sigma) added with a phosphatase inhibitor (Sigma), and protein concentration was quantified through the method of Bradford. Sodium dodecyl sulfate loading buffer (Sigma) was used to prepare equal concentrations of protein samples. Subsequently, electrophoresis of samples (20 μg protein) was performed using 8% of bis-tris-acrylamide gels (Yubo Biotech) under constant voltage. The separated protein bands were transferred onto PVDF membranes (GenScript). After incubation with freshly prepared skim milk powder (Sigma), the membranes were probed with TLR4 antibody (#K003881P, Solarbio) and GAPDH antibody (#K200057M, Solarbio) prepared according to the manufacturer′s protocol. At last, ECL-Plus reagent (Solarbio) was used for the visualization of the separated protein bands.
2.14 Statistical analysis
The statistical analyses were conducted using GraphPad Prism and results were shown as means ± standard deviations. For group comparisons, the study performed Wilcoxon−Mann−Whitney test, Student's t-tests, one-way and two-way analysis of variance or Spearman's correlation test. p < .05 indicated statistical significance.
3 RESULTS
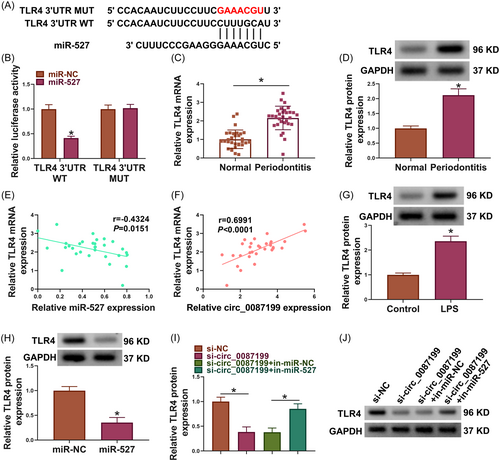
3.1 Circ_0087199 expression was upregulated in periodontal ligament tissues of periodontitis patients
Circ_0087199 is generated from the head-to-tail splicing of the exons 3−6 of the GDA gene with a spliced length of 394 nt (Figure 1A). Subsequently, the results showed that circ_0087199 expression was increased in the periodontal ligament tissues of periodontitis patients when compared with normal controls (normal volunteers with orthodontic requirements) (Figure 1B). Moreover, we found that circ_0087199 expression was increased in LPS-stimulated PDLCs in comparison with PBS-treated PDLCs (Figure 1C). RNase R treatment significantly reduced GAPDH expression but did not affect circ_0087199 expression (Figure 1D). As shown in Figure 1E, circ_0087199 was mainly expressed in the cytoplasm of PDLCs.

3.2 Circ_0087199 depletion attenuated LPS-induced apoptosis, inflammation and oxidative stress of PDLCs
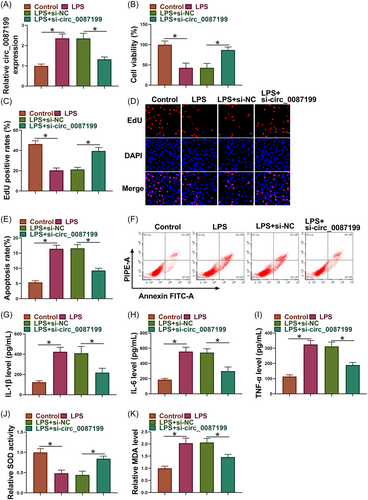
LPS treatment increased circ_0087199 expression in PDLCs, whereas the effect was relieved after circ_0087199 knockdown (Figure 2A). Subsequently, LPS treatment inhibited the viability and proliferation and induced apoptosis of PDLCs, but these effects were restored after circ_0087199 depletion (Figure 2B−F). As shown in Figure 2G−I, the increased levels of IL-1β, IL-6 and TNF-α induced by LPS treatment were rescued when circ_0087199 expression was downregulated. Comparatively, LPS treatment inhibited SOD activity and increased MDA production, however, these effects were relieved after circ_0087199 silencing (Figure 2J,K).
3.3 Circ_0087199 bound to miR-527 in PDLCs
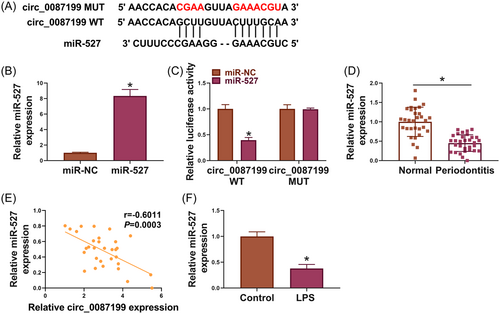
The binding sites of circ_0087199 for miR-527 are presented in Figure 3A. The results of qRT-PCR analysis indicated the high efficiency of miR-527 overexpression in PDLCs (Figure 3B). Subsequently, miR-527 mimics inhibited the luciferase activity of circ_0087199 WT but not that of circ_0087199 MUT (Figure 3C). As shown in Figure 3D and E, miR-527 expression was downregulated and negatively correlated with circ_0087199 expression in the periodontal ligament tissues of periodontitis patients. Moreover, LPS-stimulated PDLCs showed a low miR-527 expression in comparison with PBS-treated PDLCs (Figure 3F).
3.4 MiR-527 depletion relieved circ_0087199 knockdown-mediated effects in LPS-induced PDLCs
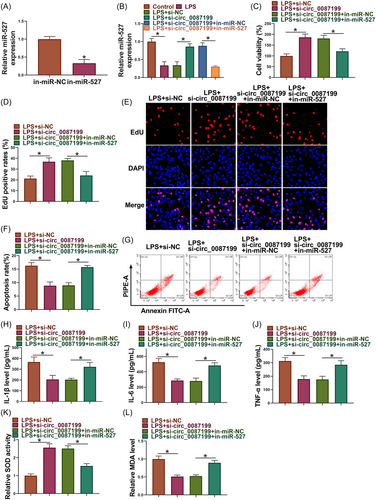
As shown in Figure 4A, the efficiency of miR-527 inhibitors in reducing miR-527 expression was high in PDLCs. Subsequently, circ_0087199 knockdown upregulated miR-527 expression in LPS-induced PDLCs, whereas the effect was restored after transfection with miR-527 inhibitors (Figure 4B). The increased cell viability and proliferation and decreased cell apoptosis by circ_0087199 knockdown were restored after transfection with miR-527 inhibitors in LPS-induced PDLCs (Figure 4C−G). Comparatively, circ_0087199 silencing reduced IL-1β, IL-6 and TNF-α levels, promoted SOD activity and inhibited MDA production in LPS-induced PDLCs, however, these effects were relieved when miR-527 expression was decreased (Figure 4H−L).
3.5 Circ_0087199 regulated TLR4 by binding to miR-527
The binding sites of miR-527 for TLR4 are shown in Figure 5A. Subsequently, miR-527 mimics inhibited the luciferase activity of TLR4 3′UTR WT but not that of TLR4 3′UTR MUT (Figure 5B). TLR4 expression at mRNA and protein levels was upregulated in the periodontal ligament tissues of periodontitis patients when compared with normal controls (Figure 5C,D). Comparatively, TLR4 expression was negatively correlated with miR-527 expression but positively with circ_0087199 expression in the periodontal ligament tissues of periodontitis patients (Figure 5E,F). LPS-stimulated PDLCs showed a high TLR4 expression in comparison with PBS-treated PDLCs (Figure 5G). As shown in Figure 5H, miR-527 overexpression significantly inhibited TLR4 protein expression in PDLCs. Further, circ_0087199 silencing reduced TLR4 protein expression, whereas the effect was relieved after transfection with miR-527 inhibitors (Figure 5I,J).
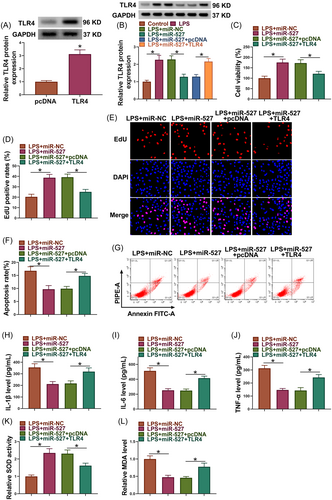
3.6 TLR4 introduction rescued miR-527 mimic-induced effects in LPS-induced PDLCs
As shown in Figure 6A, the efficiency of TLR4 overexpression was high in PDLCs. Subsequently, miR-527 mimics inhibited TLR4 protein expression in LPS-induced PDLCs, whereas the effect was restored after transfection with TLR4 overexpression plasmid (Figure 6B). The increased cell viability and proliferation and decreased cell apoptosis by miR-527 mimics were restored after TLR4 overexpression in LPS-induced PDLCs (Figure 6C−G). Comparatively, miR-527 introduction reduced IL-1β, IL-6 and TNF-α levels, promoted SOD activity and inhibited MDA production in LPS-induced PDLCs, however, these effects were relieved when TLR4 expression was increased (Figure 6H−L).
4 DISCUSSION
Periodontitis is a chronic disorder that can destruct periodontal ligament, cementum, and bone. Susceptibility to periodontitis is affected by smoking, uncontrolled diabetes mellitus, and both genetic and epigenetic factors. Emerging research indicates that epigenetic-associated ncRNAs such as circRNAs mediate periodontitis pathogenesis by promoting periodontal regeneration, regulating periodontal ligament stem cell differentiation, and regulating dental pulp stem cell differentiation.15 Identification of abnormal circRNAs in periodontitis is needed to explore novel therapeutic avenues for periodontitis. PDLCs are a class of main cell populations in diseased periodontium.16 The present work analyzed the effects of circ_0087199 on LPS-induced apoptosis, inflammation and oxidative stress of PDLCs. The results showed that circ_0087199 knockdown exerted inhibitory effects on PDLC injury induced by LPS and that miR-527/TLR4 pathway was required for circ_0087199-mediated actions in PDLCs.
Previous studies have revealed that some circRNAs such as circ_0138960 and circ_0081572 are involved in LPS-induced dysfunction of PDLCs.17, 18 Our work reported the involvement of another novel circRNA circ_0087199 in LPS-induced injury of PDLCs for the first time. Our data showed that circ_0087199 expression was upregulated in periodontal ligament tissues of periodontitis patients, which are consistent with previous results.11 LPS-treated PDLCs also showed a high circ_0087199 expression. The covalent loop structures of circRNAs ensure that they are resistant to RNase R digestion.19 Our data also confirmed that circ_0087199 had strong resistance to RNase R degradation in comparison with linear RNA (GAPDH). Additionally, circ_0087199 depletion rescued LPS-caused proliferation inhibition and apoptosis promotion of PDLCs. Moreover, LPS pretreatment-induced promoting effects on inflammation and oxidative stress of PDLCs were relieved after circ_0087199 silencing. Thus, circ_0087199 may contribute to periodontitis progression.
Thorough and extensive research has focused on miRNA function in the pathophysiology of periodontal diseases.20 Certain bacterial components isolated from the oral bacterial plaque induced abnormal miRNA expression, indicating the possible role of miRNAs in periodontitis progression.14 MiR-527, an inflammatory disease-related miRNA, was confirmed to be targeted by circ_0087199 in our work. MiR-527 is a newly identified miRNA related to progression of cancers such as gastric cancer,21 glioma22 and ovarian cancer.23 But there are almost no studies regarding its function in periodontitis. As reported in 2022, circ_0138959-mediated effect on gingival fibroblast pyroptosis involved miR-527.24 The present work revealed its roles in regulating apoptosis, inflammation and oxidative stress of PDLCs after periodontitis. Herein, miR-527 expression was downregulated in periodontal ligament tissues of periodontitis patients. Moreover, miR-527 expression was reduced after LPS exposure in PDLCs. miR-527 introduction ameliorated LPS-induced injuries of PDLCs including apoptosis, inflammation and oxidative stress. Importantly, PDLCs with low circ_0087199 and miR-527 expression showed increased apoptosis, inflammation and oxidative stress after LPS treatment in comparison with circ_0087199-deficient PDLCs. These data demonstrated the circ_0087199/miR-527 pathway mediated LPS-induced injury of PDLCs.
Toll-like receptor (TLR) is a specific transmembrane receptor in the natural immune system and can regulate acute inflammatory response and apoptosis.25 TLR4 belongs to the TLR family members and mediates inflammatory responses by binding to exogenous ligands and affecting endogenous transcription factor expression.26 TLR4 mediates tissue immune responses to periodontitis and is expressed in periodontal tissues.27, 28 The present study identified TLR4 as a target of miR-527. Additionally, TLR4 expression was increased in periodontal ligament tissues of periodontitis patients. In response to LPS, TLR4 exhibited a high expression in PDLCs. Previous work has revealed that TLR4 is associated with alveolar bone destruction in periodontitis.29 Li et al. have reported that TLR4 pathway is activated in response to LPS stimulation to regulate NF-κB pathway, an important signaling pathway in periodontitis.30, 31 Our data showed that PDLCs with high miR-527 and TLR4 expression showed increased apoptosis, inflammation and oxidative stress after LPS treatment in comparison with PDLCs with only high miR-527 expression. Importantly, circ_0087199 induced TLR4 expression by interacting with miR-527.
Collectively, circ_0087199 knockdown attenuated LPS-induced apoptosis, inflammation and oxidative stress of PDLCs by regulating the miR-527/TLR4 pathway. However, comprehensive roles of circ_0087199 in the pathophysiological processes of periodontitis should be analyzed because one circRNA usually has many target miRNAs. Additionally, in vivo data are lacking, and further investigation needs to be performed to provide in vivo evidence of circ_0087199 regulating periodontitis progression. Nevertheless, this work provides a theoretical basis for further drug administration of periodontitis.
AUTHOR CONTRIBUTIONS
Jing Wang performed the research. Rui Xiao designed the research study. Na Huo contributed essential reagents or tools. Chuan Cai analyzed the data. Yu Zhang wrote the paper. All authors have read and approved the final manuscript.
ACKNOWLEDGMENTS
This research was supported by the National Key R&D Program of China (Key Special Project for Marine Environmental Security and Sustainable Development of Coral Reefs 2022-2.1) and Military medical Science Foundation of Youth Project (21QNPY104).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.