STRipy: A graphical application for enhanced genotyping of pathogenic short tandem repeats in sequencing data
Abstract
Expansions of short tandem repeats (STRs) have been implicated as the causal variant in over 50 diseases known to date. There are several tools which can genotype STRs from high-throughput sequencing (HTS) data. However, running these tools out of the box only allows around half of the known disease-causing loci to be genotyped. Furthermore, the genotypes estimated at these loci are often underestimated with maximum lengths limited to either the read or fragment length, which is less than the pathogenic cutoff for some diseases. Although analysis tools can be customized to genotype extra loci, this requires proficiency in bioinformatics to set up, limiting their widespread usage by other researchers and clinicians. To address these issues, we have developed a new software called STRipy, which is able to target all known disease-causing STRs from HTS data. We created an intuitive graphical interface for STRipy and significantly simplified the detection of STRs expansions. Moreover, we genotyped all disease loci for over two and half thousand samples to provide population-wide distributions to assist with interpretation of results. We believe the simplicity and breadth of STRipy will increase the genotyping of STRs in sequencing data resulting in further diagnoses of rare STR diseases.
1 INTRODUCTION
Short tandem repeats (STRs) are sequences consisting of repeated, short motifs up to 6 bp long. They comprise about 3% of the human genome (Subramanian et al., 2003). Over 50 human diseases are reported to be caused by expansions in the number of repeats at specific loci in the human genome and around one-fifth of these have been discovered in the past 5 years (Depienne & Mandel, 2021). Many of the recently discovered disease-causing STRs are different to the vast majority of previous findings, as the pathogenic motif was found to be absent in the reference genome, which is based on healthy individuals. These include repeats that cause different types of familial adult myoclonic epilepsies (Corbett et al., 2019; Florian et al., 2019; Ishiura et al., 2018; Yeetong et al., 2019), spinocerebellar ataxia 37 (Seixas et al., 2017), and cerebellar ataxia neuropathy vestibular areflexia syndrome (CANVAS) (Cortese et al., 2019).
Analysis of STRs from short-read sequencing data is not straightforward due to low sequence complexity and nonuniqueness of sequences from these regions. Therefore, STR analysis has often been ignored in medical sequencing studies (Gymrek, 2017). However, advances in sequencing technology and repeat-aware software over the past decade has resulted in several bioinformatics tools built for genotyping STRs. Some tools also estimate the repeat length for alleles longer than fragment length, such as ExpansionHunter (Dolzhenko et al., 2017) and GangSTR (Mousavi et al., 2019). ExpansionHunter is a tool created specifically for targeted genotyping analysis and has been used successfully on a large-scale analysis of rare diseases, detecting alleles which are shown to be well correlated with polymerase chain reaction (PCR)-based assays (Ibanez et al., 2020). The latest version (5.0.0) of the tool uses a variant catalog containing information, which allows the estimation of STR genotypes in 29 loci that are known to cause a disease. For only two of these loci, genotypes that are longer than the sequencing fragment length can be determined. In addition, the provided variant catalog does not contain information for detecting any of the recently discovered loci where a pathogenic motif was inserted between or next to the endogenous STR.
STRs genotyping tools are command-line programs that fit well into bioinformatic pipelines. However, they are not intuitive to set up and use on their own without relevant skills, limiting their everyday application to the analysis of sequencing data. Here we introduce a new free and open-source software with graphical interface called STRipy, which is an easy-to-use program built to detect all known pathogenic STRs to date. The first aspect of STRipy is the curated database of all discovered disease-causing STRs with supporting genomic and pathogenic information. STRipy incorporates ExpansionHunter (Dolzhenko et al., 2017) into its framework as the backend genotyper and uses the database to increase the number of defined loci to genotype from 29 to 55. STRipy now includes all currently known disease-causing loci. We additionally integrated the REViewer tool (Dolzhenko et al., 2021) to visualize reads aligned to each haplotype, which also enables the visualization of any interruptions or variants in the STR locus. Finally, we added functionality to STRipy, to enable genotyping of alleles that are longer than the sequencing fragment length and also report the number of pure pathogenic motifs in reads. Aligned whole genome, whole exome or targeted Illumina sequencing files in BAM (binary alignment map) or CRAM (compressed and reference-oriented alignment map) format can be used as input to STRipy and genotyping will be performed as long as there is coverage on the targeted STR locus.
To provide the distribution of STR lengths at the population level, we genotyped 2504 individuals from the 1000 Genomes Project cohort (Auton et al., 2015; Illumina Inc., 2020). This, together with information from the literature, is provided to improve assessment of pathogenicity of genotype in a sample. The resulting online database of disease-causing STRs with population data for all loci provides a valuable resource for scientists in the field. Ultimately, STRipy aims to improve the clinical diagnoses of rare diseases by allowing the easy detection of STRs expansions in pathogenic loci.
2 DESIGN AND IMPLEMENTATION
2.1 STRipy's Client
STRipy is implemented in Python 3 and consists of two parts—Client and Server. Both can be easily installed on Linux and macOS operating system as well as Windows 10/11 through the Windows Subsystem for Linux. The Client is the main part of the software, which a user interacts with through a graphical interface. It allows the genotyping of one locus at a time to provide quick results and avoid incidental findings. The whole genotyping process is streamlined, only requiring a user to select an indexed BAM or CRAM file of aligned reads and specify either a locus (gene) to target or a disease associated with an STR expansion. The genome reference can be defined by the user or detected automatically for simplicity. STRipy's autodetection algorithm scans the sequences before and after the targeted STR locus in the analyzed sample to determine the most likely genome. If no matches are found, the user needs to select it manually.
The type of analysis can be chosen to be either “Standard” or “Extended.” Standard analysis is fast and recommended for use in the first instance, but the detected genotyping length is limited to the fragment length. Extended analysis enables genotyping of alleles longer than the fragment length. STRipy's extended analysis does the extra process to try and recover misaligned reads from off-target regions, but can result a more time-consuming analysis. Finally, a user can choose whether to run the analysis in STRipy's Server installed into their own computer/internal network (Local) or in a server running on cloud (Cloud). Upon clicking on the button to genotype the sample, a cascade of processes will be executed.
STRipy uses the catalog containing variant information such as the reference coordinate and sequence of the repeat unit required for the bioinformatics analysis. At first, STRipy's Client extracts out reads from the BAM/CRAM file overlapping the STR and a flanking region of 2 kb, by default, on each side of the STR locus (Figure 1). When using the “Standard” analysis, only flanking reads from the STR region and fully repeated reads with a pair in the extracted region will be included into the analysis file. When using the “Extended” analysis, as well as extracting out mates of the reads aligned next to the STR locus, STRipy takes the mapped locations of these repeat reads to identify other fully repeat reads made of the pathogenic repeat (Level 1 [L1] off-target regions). If they exist, then these reads will be extracted out as well with their mates, some of which can be aligned to another location (Level 2 [L2] off-target region). The resulting analysis ready file for this “Extended” analysis includes reads from the region surrounding the reference STR locus and reads found from L1 and L2 off-target regions. These off-target regions are provided to ExpansionHunter to extend the genotyping estimation above the fragment length.
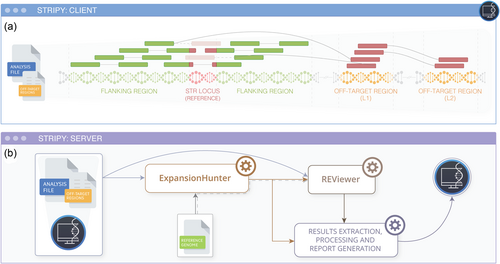
Additionally, STRipy determines the number of pathogenic motifs in reads. This can be useful when the allele contains interruptions or is made of repeats different from the endogenous ones. The presence or lack of the pathogenic repeat unit is critical information to the user when analyzing replaced and nested type of repeats as ExpansionHunter counts similar repeat units into the length of the reported genotype even when there is no pathogenic motif present. Finally, a BAM file with an anonymized file name is created, containing the extracted reads, which is then indexed and forwarded to STRipy's Server for genotyping.
2.2 STRipy's Server
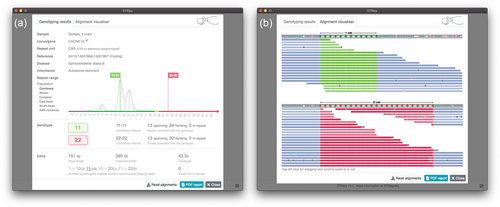
STRipy's Server listens for inputs from the Client and performs several tasks after receiving the data. First, the received BAM file will be genotyped by ExpansionHunter (Dolzhenko et al., 2017) using the reference coordinate and repeat unit which was acquired from the STRipy database. In the case of an “Extended” analysis, off-target regions found by STRipy's Client will be forwarded to ExpansionHunter to enable genotyping of long alleles (where the repeat region is longer than the fragment length). This approach is different from using ExpansionHunter outside of STRipy, as we use the data to determine off-target regions for each sample individually, in contrast to using predefined regions for all samples. After genotyping, ExpansionHunter's output files will be analyzed by the tool REViewer (Dolzhenko et al., 2021), which creates read alignment visualizations for the STR region. Read visualization is an important part of the results section for two reasons. Firstly, it allows visual assessment of how well the genotype is supported by reads aligned to the locus. Secondly, it shows the most likely sequence for both alleles, including the presence of interruptions within repeats, which can be clinically important, for example, by impacting the timing of disease onset (Wright et al., 2019). In the end, genotyping results from ExpansionHunter along with the visualizations of read alignments are matched with the pathogenicity ranges acquired from the literature. The Client then presents the genotyping results, accompanied by repeat-specific information obtained from literature, as well as read visualizations and the population-wide data we have generated, to improve assessment of the results. This report is returned to the user in the graphical interface and as a PDF (Figure 2).
The server requires Python 3 with svglib, regex, pyramid, and reportlab library, and needs to have access to the reference genome(s) as well as to Samtools (Li et al., 2009), ExpansionHunter, and REViewer executables. To further simplify the process of genotyping STRs, we provide a free STRipy's Server (Cloud) from our end where all the tools and main reference genomes are provided. The Cloud has been configured for maximum privacy and it does not store user-identifying details or any information about the sample (including sequencing data and genotyping results). By using the Cloud, a user does not have to set up a server on their own. This makes genotyping STRs an exceptionally easy process, as there is no installation or set-up when using the compiled STRipy's Client which contains Python 3, as well as all the required libraries (pysam, pywebview, requests, numpy, and regex).
2.3 STRipy's STR database
We created and made available the most comprehensive and up-to-date STR disease database for use by STRipy and for more general purposes. This database contains variant information for each disease locus assisting with bioinformatics analysis and results interpretation. We performed an in-depth search of the literature to determine all disease-causing STRs loci reported so far and manually adjusted all reference coordinates, as well as repeat unit sequences for different genome versions (Table S1–S3). At the time of writing, we determined 55 loci in 52 genes where repeat expansions are reported to be causative for 58 diseases.
We devised a strategy to classify our list of loci into three groups based on the repeat type: standard (34 loci), imperfect GCNs (12 loci), and replaced/nested (9 loci). Standard repeats are those where a specific repeat unit (e.g., CAG or CCG) is found in healthy individuals and its expansion becomes pathogenic at larger numbers of repeats (Table S1). Imperfect GCN-type repeats refer to sequences that encode for the amino acid alanine, but the sequence can be composed of different, but synonymous, repeat units—either GCA, GCG, GCC, or GCT (Table S2). Lastly, replaced/nested type repeats are where the pathogenic motif is not present in the reference genome and usually not found in healthy individuals (Table S3). Out of the nine diseases discovered so far, only one (CANVAS) is caused by replaced type repeats. Here, the repeat unit found in healthy individuals (AAAAG) is replaced with a sequence composed of a different motif (either AAGGG or ACAGG) in diseased individuals. All the other diseases in this group are of the nested type where a repeat of the pathogenic motif (such as TTTCA) is inserted between or next to the nonpathogenic stretch of endogenous repeats (such as TTTTA).
Finally, we used ExpansionHunter to genotype all 55 loci in 2504 samples from the DRAGEN reanalysis of 1000 Genomes Project data set (Illumina Inc., 2020) to obtain population-wide distributions. This data set includes samples in five superpopulation and in 26 populations, which a user can view separately or download together. We believe utilization of the population information can help a user to determine whether the genotyping results could be clinically important.
We have made this curated set of disease-causing STRs expansions available as a public resource, which will be maintained and updated with new disease-causing STR expansions as they are discovered and reported.
3 RESULTS
To validate and assess STRipy's performance, we conducted a series of simulations where we validated one locus in each gene. First of all, we simulated heterozygous samples by using ART next-generation sequencing read simulator tool (Huang et al., 2012). One allele was set to be 60 bp in length and we varied the length of the other allele in the range of 60–2100 bp by adding one motif length for each simulation. This includes a large range of repeat lengths and includes the pathogenic cutoff in ~90% of loci. In the same manner, we simulated rarer, expanded homozygous alleles with both alleles the same length ranging from 60 to 2100 bp. These simulations were conducted for each of the 52 genes/loci to examine genotyping accuracy with increasing length of repeats (further details of the methods in Supporting Information S1 and simulated allele formulas for replaced/nested type of repeats in Table S4). Once all simulated data sets were generated (63,580 samples), we aligned the simulated reads to the hg38 reference genome using BWA-MEM (Li, 2013) and genotyped all samples with STRipy using the latest release of the ExpansionHunter at the time of the analysis (v4.0.2). To estimate the accuracy of calls across simulations we calculated root mean square error (RMSE) for each locus across the different lengths of simulations to determine the genotyping bias (Figure 3).
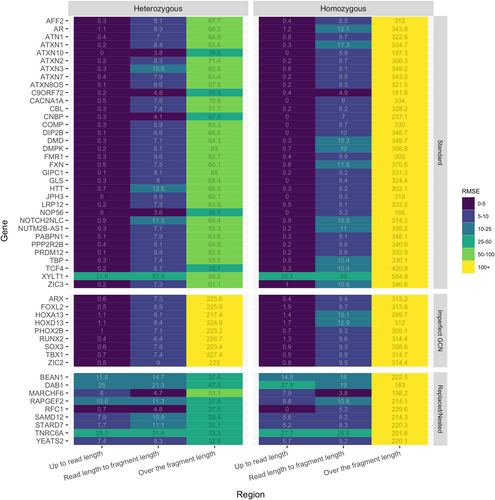
Results were obtained both for homozygous and heterozygous samples at three different STR length ranges considering whether the simulated repeat length is less than the simulated read length of 150 bp, is between the read and the fragment length (mean of 450 bp), or is longer than the fragment length (up to the simulated maximum of 2100 bp). Overall, very low RMSEs were seen across all standard and imperfect GCN type of repeats “up to read length” in both heterozygous and homozygous samples (median RMSE 0.25 for heterozygous and 0.75 for homozygous samples). We also saw low error rates for genotyping these types of STRs between the read and fragment length where the median RMSE for standard repeats was 8.60 and for imperfect GCNs 8.56. Collectively, these results show very good estimation of alleles up to the fragment length.
We demonstrated the ability of STRipy to genotype long heterozygous alleles in standard type repeats at lengths over the average fragment length of 450 bp and up to 2100 bp, resulting in the median RMSE of 63.45. An example can be seen on Figure 4B showing that STRipy is able to find the vast majority of misaligned reads and provide their location to ExpansionHunter as off-target regions, to improve genotyping of long alleles. However, we did see a slight underestimation of long alleles, which could be due to some fully repeat reads containing more sequencing errors than allowed by STRipy's algorithm and therefore not extracting them out. Genotyping the same locus using ExpansionHunter with the default variant catalog shows the inability to genotype long alleles without using our extracted misaligned fully repeated reads in off-target regions (Figure 4A). For the imperfect GCN repeats group, the median RMSE was higher, 224.97 (an example in Figure 4D). It is important to note that we simulated an extreme case of this type of repeats where the STR locus included four different alanine coding triples in random order, which potentially resulted in many unaligned reads of fully repeated GCN sequences, and therefore limiting the genotyping of long alleles. We expect more accurate estimation of genotypes in real samples where the expanded alleles have more uniform sequence and align better to the genome.
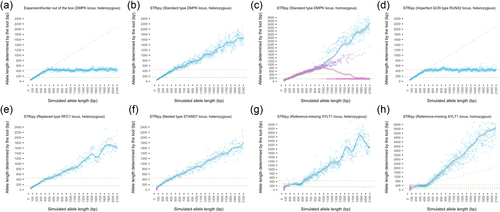
We saw significantly less accurate genotypes for homozygous samples containing alleles longer than the fragment length (median RMSE of 332.06 for standard and 313.20 for imperfect GCN types). Upon further investigation, we discovered this to be a result of ExpansionHunter algorithm where, from about 700–800 bp, the two allele length estimates start to diverge producing heterozygous estimates with one allele an underestimate and one an overestimate (Figure 4C). As we genotyped our samples well beyond that range, the resulting error rate was high. Estimation of homozygous alleles over ~700 bp is likely to be error prone with ExpansionHunter v4.0.2 and therefore also in STRipy until this gets resolved in ExpansionHunter.
Regarding the replaced type of repeats, we saw similar results for RFC1 (the only replaced type sample) to the results previously described for standard type repeats (Figure 4E). However, for the nested type of repeats where a new repeat motif is added to the repeat already documented in the reference, the RMSE in the “up to the fragment length” section was higher due to the presence of both repeats, which ExpansionHunter cannot distinguish and therefore reported a genotype of both repeat lengths combined (an example in Figure 4F). As we simulated variable numbers of endogenous repeats before and/or after the pathogenic locus, STRipy returned a genotype longer than the simulated insertion, which is clearly visible in the “up to read length” range. We developed a feature in STRipy to tackle this issue by counting the number of pathogenic repeat motifs in all spanning/flanking and fully repeated reads. If no pathogenic repeats were found in any of the spanning/flanking or fully repeated reads then this is reported on the results page, which indicates the presence of only endogenous repeat sequences. In the vast majority of simulations, STRipy found fully repeated reads containing the pathogenic motif in all samples when the inserted repeated sequence exceeded read length and reported the number of reads to user as an indication of true pathogenic expansions.
Expansions in one standard type of locus, XYLT1, are in a region that is not included in the human reference genome hg19 or hg38 (LaCroix et al., 2019). Homozygous expansions in this repeat were found to cause Desbuquois dysplasia type 2. The reference genome excludes 238 bp of which 42 bp are 14 repeats of the GCC motif with a GCT interruption after the fifth repeat. For this locus, we simulated samples with the 238 bp sequence inserted back into the XYLT1 promoter region, as well as varying the length of the STR expansion similar to other samples. The reference coordinates were specified at a reference GGC-repeated region (GCC on the positive strand) in the XYLT1 promoter, next to the missing region. Without realignment to a reference genome containing the missing region, it is difficult to accurately estimate the repeat length in the locus. This is illustrated by the high RMSE of the locus (Figure 3). Despite the lower accuracy in genotyping, STRipy was still able to recover reads and determine expansions over the fragment length in the locus (Figure 4G,H). Homozygous sample expansions were reported as heterozygous genotypes which were significantly overestimated for expansions longer than the fragment length (Figure 4H). Therefore, although it is not accurate to genotype the XYLT1 locus, we showed that STRipy can report expansions in the pathogenic range for one allele in samples with disease-causing expansions.
During our analysis, we often observed that ExpansionHunter-detected pathogenic genotypes in the first and second tract of the HOXA13 gene and, in some cases, also in tracts of the ARX gene. These are stable polyalanine loci with low mutation rates and there are no previous reports that any of these loci have expansions longer than 36 repeats. This is much shorter than alleles we reported in the population data that were up to 80 repeats long. In that data set of 2504 samples (5008 alleles), the pathogenic length reported at a rate of about 0.0002, 0.0325, 0.0202, 0.0343, and 0.0767 for ARX_1, ARX_2, HOXA13_1, HOXA13_2 and HOXA13_3 locus, respectively. The longest expansion of alanines in gnomAD v3.1.1 database (Karczewski et al., 2020) for any of these loci is no more than 10 repeats. The average frequency for a pathogenic expansion in gnomAD is very low (0.0001230 for ARX_1, 0.0000096 for ARX_2, 0.0000207 for HOXA13_1, 0.0000137 for HOXA13_2 and 0.0000072 for HOXA13_3), indicating that STRipy's expansions are false positives. In addition, we manually examined those samples and generally saw a large number of mismatched bases and low coverage of the expanded alleles, indicating improperly aligned and genotyped alleles.
As this was observed only for these specific loci, we assume this is due to the proximity of the two tracts of the same, imperfect GCN-type motifs. For example, HOXA13_1 and HOXA13_2 both have GCN repeats 63 bp apart, and ARX_1 and ARX_2 have the same repeats with an 84 bp gap. In STRipy, we resolved this issue by limiting the region for extracting reads from these loci to 60 bp (optional setting), which then only extracts out spanning reads in this locus that are sufficient for genotyping such loci with a small pathogenic cutoff. This results in only using the local reads to infer the genotype, while the repeats in the adjacent tract are not included. This now leads to more accurate results in STRipy (genotyping results of HOXA13 loci in 10 biological samples are shown in Table S5).
Finally, STRipy was applied to a set of whole genome sequenced (WGS) samples that was previously used to validate the genotyping tool STRetch (Dashnow et al., 2018) and which contains known pathogenic expansions for nine samples plus one in the intermediate range. We firstly analyzed the pathogenic locus in all 10 samples (Table 1) in “Standard” mode, for which 7 were determined to be in the pathogenic range (5 of them with an estimated allele length at least as long as measured by PCR), as well as the 1 in the intermediate range. For the samples where the allele was less than the PCR length (except Number 5), a notification was displayed to the user about a potential presence of a long allele with the recommendation to use “Extended” analysis. When using the “Extended” analysis, the estimated allele length for all affected individuals, except Number 5, was in the pathogenic range and for six of them at least the measured PCR value. For the other two, the PCR genotype was in the confidence interval and also close to the estimated one (six repeat difference). Results and visualizations of sample Number 4 are also displayed as an example on the Figure 1.
| STRipy (standard mode) | STRipy (extended mode) | |||||||
|---|---|---|---|---|---|---|---|---|
| # | Gene | Pathogenic cutoff | Measured PCR | Genotype | CI | Genotype | CI | |
| 1 | ATXN1 | 39 | 51 | 30/59 | 30-30/52-69 | 30/59 | 30-30/52-69 | |
| 2 | ATXN1 | 39 | 29/32 | 29/34 | 29-29/34-34 | 29/34 | 29-29/34-34 | |
| 3 | ATXN3 | 56 | 73 | 24/67 | 24-24/56-82 | 24/67 | 24-24/57-83 | |
| 4 | CACNA1A | 21 | 22 | 11/22 | 11-11/22-22 | 11/22 | 11-11/22-22 | |
| 5 | AR | 40 | 41 | 36/36 | 27-43/35-52 | 36/36 | 28-44/35-53 | |
| 6 | C9orf72 | 31 | >50 | 12/54 | 12-12/41-72 | 12/94 | 12-12/72-127 | |
| 7 | CNBP | 55 | >75 | 15/44 | 15-15/41-62 | 15/69 | 15-15/69-127 | |
| 8 | DMPK | 50 | >150 | 5/129 | 5-5/98-154 | 5/342 | 5-5/365-407 | |
| 9 | AR | 40 | 47 | 50/56 | 38-64/50-81 | 50/56 | 38-64/50-81 | |
| 10 | FXN | 70 | ~850 | 115/115 | 62-123/80-156 | 50/871 | 49-89/539-966 | |
- Abbreviation: CI, confidence interval.
Additionally, we measured the time of analysis for all biological samples and calculated the mean with SD by using both types of analysis (Table S6). When using the “Standard” mode, the time of analysis (from clicking the genotyping button to receiving results from the server) was between 4.1 and 20.4 s (mean 10.4, SD 4.63). The “Extended” analysis had high variation and analyses took from 4.6 and up to 281.8 s (mean 77.1, SD 107.26). The time can depend on numerous factors, including but not limited to the number and size of off-target regions (in case of the “Extended” analysis), coverage and type of the sequencing file, computer's processing power and internet speed when using the Cloud.
4 DISCUSSION
Here we present STRipy, a software and database that can be easily used to genotype any known disease-causing STRs locus. STRipy works very well to estimate the repeat length in all standard and imperfect GCN type of repeats that are present in the reference genome up to the read length. For standard type repeats genotyping for accurate above up to the fragment length. This enables accurate genotyping of all alleles in the pathogenic range for both standard and imperfect GCN types as the pathogenic range for all GCN type of repeats found so far have not exceeded 36 repeats or 108 bp which is below modern read lengths of 150 bp.
STRipy's backend uses ExpansionHunter as the genotyping tool and we identified three issues during our study. Firstly, ExpansionHunter exhibits difficulties in correctly genotyping long homozygous alleles above a certain threshold, in our case around 700–800 bp. However, now this has been identified, the issue can be mitigated by further manual investigation of the sequencing data to determine the diagnosis of a patient with such long alleles. Out of all the known diseases, this could return an inaccurate genotype (where one allele is not in the pathogenic range) for two diseases that are caused by long biallelic expansions exceeding 600 bp: CANVAS and often Friedreich's ataxia. In addition, only one expanded allele is reported when analyzing homozygous samples for the expansions in XYLT1 locus where the STR region is missing in the reference. Despite that, a user will be informed of one very long allele which will indicate the need for a follow up analysis. Once this issue is resolved in the ExpansionHunter software, it will be fixed automatically in STRipy without the need for changing the code.
Secondly, we observed that the genotyping of nested repeats has higher error rates. This is due to ExpansionHunter's inability to differentiate between the pathogenic and endogenous repeat units. For this reason, a genotype is reported as the sum of these repeats. However, STRipy is designed to report the number of pathogenic repeat units found in spanning/flanking reads and the number of reads fully made of the pathogenic repeat, which should be taken into account when genotyping these loci. The presence of pathogenic repeat units can also be confirmed when examining the provided read visualizations. Moreover, read visualizations provide information about the sequence in the STR locus, including any interruptions or nucleotide variations that are present, making an easy visual way for user to understand the STR locus.
Finally, we improved genotyping of alleles longer than the fragment length by determining off-target regions for misaligned reads from the data itself. However, it is important to note the possibility of observing an overestimated genotype if there are more loci present in the sample, which contain long uninterrupted repeats with the same motif as the targeted one, resulting in fully repeated reads, which are mistakenly used to estimate a genotype.
Here we used simulations to evaluate STRipy and, additionally, we genotyped nine different WGS samples with disease-causing expansions. Diseases caused by STR expansions are rare and some of them have only been found in single families or even individuals, which makes it nearly impossible to obtain sequencing data to validate all disease loci using real biological samples. Realistically, we can validate all loci only by using simulations and this is the main limitation of our validation. Data from real sequenced samples might be noisy and more complex to analyze than simulated samples. However, we created a large, robust, and comprehensive simulation data set to more closely resemble real life data by introducing variability in the fragment size, simulated reads with natural occurring sequencing errors and created alleles for replaced/nested type of repeats that match with biological findings.
STRipy fills a specific niche in the field and is meant to serve as the first-line screening tool for STR diseases that is available to the wider community. STRipy allows the targeting of one locus in one sample at the moment and, therefore, it is more a complementary tool for ExpansionHunter and other similar tools developed, which can be incorporated into bioinformatics pipelines and used in large scale multiloci analysis. We have also provided a tool for creating ExpansionHunter's variant catalog to assist users with such large-scale analyses, which contains all loci described in this study.
STRipy and the STRs database is available at https://stripy.org. As new disease-causing STR expansions are discovered these will be added to STRipy's database and genotyping functionality.
ACKNOWLEDGMENT
Open access publishing facilitated by The University of Melbourne, as part of the Wiley - The University of Melbourne agreement via the Council of Australian University Librarians.
CONFLICTS OF INTEREST
Egor Dolzhenko is an employee of Illumina, Inc., a public company that develops and markets systems for genetic analysis.
Open Research
DATA AVAILABILITY STATEMENT
The software is available on a GitLab repository at https://gitlab.com/andreassh/stripy-client (STRipy's Client) and https://gitlab.com/andreassh/stripy-server (STRipy's Server). STRipy's version 1.1 source code is also archived on Zenodo (https://10.5281/zenodo.5709228). All other code and data used in this study is available on Zenodo (https://10.5281/zenodo.5745740). STRipy's database is available at https://stripy.org/database. The true positive biological whole genome samples are available from the Sequence Read Archive under the accession number SRP148723 (individual samples accessions are from SRX4114164 to SRX4114173).




