LINE- and Alu-containing genomic instability hotspot at 16q24.1 associated with recurrent and nonrecurrent CNV deletions causative for ACDMPV
Communicating by Haig H. Kazazian
Abstract
Transposable elements modify human genome by inserting into new loci or by mediating homology-, microhomology-, or homeology-driven DNA recombination or repair, resulting in genomic structural variation. Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is a rare lethal neonatal developmental lung disorder caused by point mutations or copy-number variant (CNV) deletions of FOXF1 or its distant tissue-specific enhancer. Eighty-five percent of 45 ACDMPV-causative CNV deletions, of which junctions have been sequenced, had at least one of their two breakpoints located in a retrotransposon, with more than half of them being Alu elements. We describe a novel ∼35 kb-large genomic instability hotspot at 16q24.1, involving two evolutionarily young LINE-1 (L1) elements, L1PA2 and L1PA3, flanking AluY, two AluSx, AluSx1, and AluJr elements. The occurrence of L1s at this location coincided with the branching out of the Homo-Pan-Gorilla clade, and was preceded by the insertion of AluSx, AluSx1, and AluJr. Our data show that, in addition to mediating recurrent CNVs, L1 and Alu retrotransposons can predispose the human genome to formation of variably sized CNVs, both of clinical and evolutionary relevance. Nonetheless, epigenetic or other genomic features of this locus might also contribute to its increased instability.
1 INTRODUCTION
Approximately 45% of the human genome is composed of transposable elements (TEs), a small fraction of which is still capable of undergoing transposition in both germline and somatic cells (Beck, Garcia-Perez, Badge, & Moran, 2011; Boissinot & Sookdeo 2016; de Koning, Gu, Castoe, Batzer, & Pollock, 2011; Furano, 2000; Helman et al., 2014; Ivancevic, Kortschak, Bertozzi, & Adelson, 2016; Jurka, 2000; Lander et al., 2001; Lee et al., 2012). The presence of TEs has profound implications as they contribute to genome evolution and disease (Beck et al., 2010; Callinan & Batzer 2006; Gogvadze & Buzdin 2009; Hancks & Kazazian 2016; Iskow et al., 2010; Kazazian & Moran 2017; Richardson et al., 2015).
In addition to insertional mutagenesis and nonpathogenic intra- and interindividual variation, mobile elements can act as substrates for homology-driven rearrangements. Similar to low-copy repeats or segmental duplications, LINE-1 (L1) and endogenous retroviral elements (ERVs) can predispose the genome to copy-number variant (CNV) deletions and reciprocal duplications via nonallelic homologous recombination (NAHR; Belancion, Deininger, & Roy-Engel, 2009; Boone et al., 2014; Burwinkel & Kilimann 1998; Campbell et al., 2014; Gilbert, Lutz, Morrish, & Moran, 2005; Hedges & Deininger 2007; Hehir-Kwa et al., 2016; Higashimoto et al., 2013; Kohmoto et al., 2017; Lupski 2010; Quadri et al., 2015; Startek et al., 2015; Szafranski et al., 2016; Temtamy et al., 2008; Vissers et al., 2009). Other rearrangements mediated by L1s and ERVs include translocations (Buysse et al., 2008; Robberecht et al., 2013), insertions (Gu et al., 2016), inversions (Kidd et al., 2010), and complex genomic rearrangements (Gu et al., 2015; Liu et al., 2011). L1s, ERVs, and Alu elements also predispose the genome to structural variants via DNA break repair- or replication-associated processes (Carvalho & Lupski 2016). Due to the high copy-number of retrotransposons, CNVs mediated by them remain challenging for detection using chromosomal microarray analysis or next generation sequencing that rely on sequence uniqueness to identify assay results by specific genomic coordinates (Hehir-Kwa et al., 2016; Thung et al., 2014).
Recently, we have compiled 49 CNV deletions in the FOXF1 locus causative for alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV; MIM# 265380; Szafranski et al., 2016). ACDMPV is a lethal neonatal lung developmental disorder characterized by severe respiratory failure and refractory pulmonary hypertension (Bishop, Stankiewicz, & Steinhorn, 2011; Langston, 1991). The vast majority of patients with ACDMPV had point mutations or CNV deletions in FOXF1 or its distant upstream enhancer on 16q24.1 (Sen et al., 2013; Stankiewicz et al., 2009; Szafranski et al. 2013; Szafranski et al., 2016). Interestingly, over three-fourths of the ACDMPV causative deletions, for which breakpoints were sequenced, involved retrotransposons; in 30% of those cases, L1 was present at least at one of the CNV two breakpoints, and half of the deletions were Alu-mediated.
Here we describe a novel genomic instability hotspot at 16q24.1, featuring L1 and Alu elements located at the distal edge of the FOXF1 enhancer region, and show that it is involved in formation of same-sized and variably sized pathogenic and benign CNVs.
2 METHODS
2.1 Human subjects
ACDMPV patients and their parents were recruited and tissue specimens were collected after obtaining informed consents, following protocols approved by the IRB for Human Subject Research at Baylor College of Medicine (H-8712).
2.2 Lung autopsy and biopsy
Histopathological initial evaluations and subsequent verification were done using formalin-fixed paraffin-embedded (FFPE) tissue specimens from lung biopsies or autopsies stained with hematoxylin and eosin.
2.3 DNA isolation
DNA was extracted from peripheral blood or FFPE lung tissue using Gentra Purgene Blood Kit (Qiagen, Germantown, MD) and DNeasy Blood and Tissue Kit (Qiagen), respectively.
2.4 Array comparative genomic hybridization
CNV deletions were identified by comparative genomic hybridization (CGH) using custom-designed high-resolution, 16q24.1 region-specific oligonucleotide microarrays (4 × 180K; Agilent Technologies, Santa Clara, CA). Array CGH (aCGH) was performed according to the Agilent Technologies aCGH protocol v3.5.
2.5 Sequencing of deletion breakpoints
Deletion junctions were amplified by long-range polymerase chain reaction (PCR) using LA Taq DNA polymerase (TaKaRa Bio, Madison, WI). Cycling conditions were 94°C for 30 s and 68°C for 7 min, repeated 30 times. Primers were designed with Primer3 (https://frodo.wi.mit.edu/primer3) using up to 10 kb-large breakpoint-containing regions determined by aCGH. PCR products were treated with ExoSAP-IT (USB, Cleveland, OH) and directly Sanger sequenced. Sequences were assembled using Sequencher v.4.8 (GeneCodes, Ann Arbor, MI) and the reference human genome version GRCh37/hg19 (https://genome.ucsc.edu).
2.6 Parental origin of deletions
Parental origin of the deletions was determined using informative microsatellites or single nucleotide polymorphism mapping to the deleted genomic interval.
2.7 Distribution of the recombination-associated motif along 16q24.1
The copy number of the 7-mer 5′-CCTCCCT-3′ motif along the 16q24.1 region was compared with the expected copy number of this motif estimated by simulation assuming its uniform distribution. We checked 1,000 randomly sampled regions equal in length to the 16q24.1 region and calculated the number of the recombination-associated motifs along the analyzed region. We justified the evidence of enrichment of the 7-mer recombination motif by checking the frequency of several randomly chosen 7-mers.
2.8 In silico phylogenetic analyses of ACDMPV-linked L1PA2 and L1PA3 and Alu elements
BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was conducted for homologs of L1PA3 (chr16:86,266,902-86,272,916) and L1PA2 (chr16:86,295,780-86,301,803) on chromosome 16. Sequences with length cutoff of 5 kb and identity cutoff of 96% were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). Phylogenetic reconstruction was then performed using the maximum-likelihood method implemented in the R "phangorn" package (https://cran.r-project.org/web/packages/phangorn/phangorn.pdf) with GTR + Γ + I model of evolution (the general time reversible model with corrections for invariant characters and gamma-distributed rate heterogeneity). The tree was rooted in the L1PA4 consensus sequence (Khan, Smit, & Boissinot, 2006). The nonhuman primate evolution of Alu elements in the described locus was reconstructed by sequence comparison.
2.9 PCR analyses of syntenic genomic regions in nonhuman primates
Experimental verification of genome integration times for L1PA2 and L1PA3 at 16q24.1 was done by determining the presence of their orthologs in syntenic chromosomal regions in chimp, gorilla, orangutan, and macaque by long-range PCR. Primers used for amplifications were designed from unique sequences flanking L1 elements of interest at locations syntenic for human 16q24.1.
3 RESULTS
3.1 A novel LINE and Alu genomic instability hotspot at 16q24.1
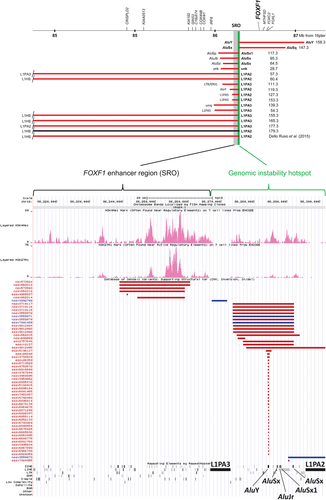
In addition to 12 previously reported CNV deletions with one breakpoint mapping at the distal edge or within the FOXF1 upstream enhancer region (Dello Russo et al., 2015; Szafranski et al., 2016), using aCGH and Sanger sequencing, we have now identified eight novel 16q24.1 deletions. The distal breakpoints of six deletions map within either L1PA2 (chr16:86,295,780-86,301,803) (pt 153.3) or L1PA3 (chr16:86,266,902-86,272,916) (pts 54.3, 155.3, 165.3, 177.3, and 179.3). These two full-length L1s are located ∼22.9 kb apart, are directly-oriented, contain PolII promoters at their 5′ end (Figure 1; Table 1; Supporting Information, Figure S1), and both are included in the L1Base2 database of ∼13,000 full-length FLn1-L1s; https://L1base.charite.de (Penzkofer et al., 2017).
| Repetitive element containing breakpoint | ||||||
|---|---|---|---|---|---|---|
| ACDMPV pt | Deletion coordinates | Proximal | Distal | Identity between LINEs or Alus (%) | Microhomology (bp) | Proposed mechanism of CNV deletion formationa |
| 28.7 | ∼chr16:86,140,499-86,285,499 | unk | unk | unk | unk | unk |
| 64.5 | chr16:86,147,527/566-86,287,120/159 | AluSz | AluSx | 84 | 38 | MMBIR, MMEJ, or SSA |
| 95.3 | chr16:86,118,131/141-86,287,054/064 | AluJb | AluSx | 75 | 9 | MMBIR or MMEJ |
| 117.3 | chr16:86,055,159/200-86,288,226/268 | AluSp | AluSx1 | 87 | 41 | MMBIR, MMEJ, or SSA |
| 147.3 | chr16:86,287,188/199-86,848,466/477 | AluSx | AluSq | 82 | 10 | MMBIR or MMEJ |
| 158.3 | chr16:86,284,317/617-87,137,455/746 | AluY | AluY | 89 | unk | MMBIR, MMEJ, or SSA |
| 54.3 | chr16:85,910,504/580-86,271,634/710 | L1PA5 (1.7 kb) | L1PA3 | 93 | 75 | NAHR |
| 57.3 | chr16:82,014,639/716-86,300,403/481 | L1PA3 (2.6 kb) | L1PA2 | 97 | 77 | NAHR |
| 60.4 | chr16:83,673,382/476-86,298,284/378 | L1HS | L1PA2 | 97 | 93 | NAHR |
| 111.3 | chr16:86,077,955/958-86,271,915/918 | LTR/ERVL | L1PA3 | – | 2 | NHEJ, MMEJ, or MMBIR |
| 119.3 | chr16:86,148,250-86,301,591 | AluY | L1PA2 | – | 7 bp insertion at the deletion junction | NHEJ |
| 127.3 | chr16:86,209,157/194-86,301,558/595 | L1PA5 (0.6 kb) | L1PA2 | 91 | 36 | NAHR |
| 139.3 | chr16:85,877,831-86,271,338 | simple repeat (TTCC)n | L1PA3 | – | 0 | NHEJ |
| 153.3 | chr16:86,208,967/995-86,301,369/397 | L1PA5 (0.6 kb) | L1PA2 | 91 | 27 | NAHR |
| 155.3 | chr16:84,491,194/238-86,271,998/272,042 | L1HS (2.1 kb) | L1PA3 | 97 | 43 | NAHR |
| 165.3 | chr16:83,672,829/882-86,268,857/910 | L1HS | L1PA3 | 97 | 52 | NAHR |
| 177.3 | chr16:82,174,710/852-86,268,760/909 | L1PA2 | L1PA3 | 96 | 149 | NAHR |
| 179.3 | chr16:83,671,523/574-86,296,427/478 | L1HS | L1PA2 | 97 | 51 | NAHR |
| Dello Russo et al., 2015 | ∼chr16:83,676,990-86,292,585 | unk | unk | unk | unk | unk |
- a MMBIR, microhomology-mediated break-induced replication; MMEJ, microhomology-mediated end joining; NAHR, nonallelic homologous recombination; NHEJ, nonhomologous end joining; SSA, single strand annealing; unk, unknown.
In total, we have sequenced 12 ACDMPV CNV deletions with their distal breakpoints located within 16q24.1 L1PA2 (six) or L1PA3 (six), delimiting one side of the FOXF1 upstream enhancer region (Figure 1). In nine of these 12 cases, the proximal breakpoint maps to a directly-oriented full-length or incomplete L1, exhibiting 91–97% sequence identity with L1 harboring the distal breakpoint and displaying 27–149 bp microhomology at the deletion junction site. In the three remaining cases, the proximal breakpoint is located within nonhomologous repetitive sequence (AluY, LTR/ERVL, or a simple repeat [TTCC]n) with 2 bp or no microhomology (Szafranski et al., 2014, 2016).
Interestingly, we have found that L1 and Alu content in the FOXF1 locus is significantly lower than that estimated for the entire genome (Supporting Information, Table S1). We have next inquired whether distribution of the breakpoints along L1 sequences is random or it correlates with the presence of some DNA structural features. We have found that breakpoints of four CNV deletions whose proximal breakpoint L1 element was complete (pts 60.4, 165.3, 177.3, and 179.3) map in 5′ portion of the L1, whereas breakpoints of deletions with proximal breakpoint mapping to incomplete L1 (pts 54.3, 57.3, 127.3, 153.3, and 155.3) or non-L1 sequence (pts 111.3, 119.3, and 139.3) clustered within 3′ one-third portion of the L1PA2 or L1PA3 (Figure 2a). To shed more light on structural features within L1PA2 and L1PA3 that might be causatively linked to the observed nonrandom distribution of DNA breakpoints along L1 sequence and L1's susceptibility to DNA breaks in general, locations of deletion breakpoints were analyzed in the context of GC content (https://www.biologicscorp.com/tools/GCContent), GC skewness (https://stothard.afns.ualberta.ca/cgview_server) (Grigoriev, 1998), potential to form palindromic structures (Grechishnikova & Poptsova 2016), and the presence of homologous recombination-associated PRDM9-binding 7-mer 5′-CCTCCCT-3′ or degenerate 13-mer 5′-CCNCCNTNNCCNC-3′motif (Billings et al., 2013; Myers, Freeman, Auton, Donnelly, & McVean, 2008). The average GC content around sequenced breakpoints (regions of microhomology or, in its absence, those flanking breakpoints by 20 bp on each side) is 39% (SD ± 2%), thus similar to overall 42% GC content of each of these two L1PAs (Figure 2a). We have also identified a negative GC composition bias in both L1s. We have not found any correlation between the location of the L1 breakpoints and the conserved stem-loops. Interestingly, the L1PA2 and L1PA3 breakpoints map within 1.6 kb (SD ± 0.5 kb, n = 10) of a 7-mer, 5′-CCTCCCT-3′of the recombination-associated motif (chr16:86,299,271-86,299,277 and chr16:86,270,389-86,270,395, respectively). This motif is also located 121 bp upstream of L1PA2 and in opposite orientation 236 bp downstream of L1PA3. In total, seven copies of 5′-CCTCCCT-3′ are located between the two L1s. We have also found an enrichment of the 7-mer recombination motif in the entire 16q24.1 (P = 0.004; Supporting Information, Figure S2).
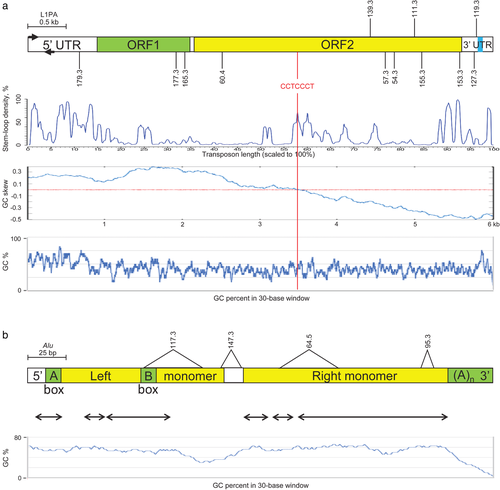
One of the two CNV deletion breakpoints in four previously reported ACDMPV patients (28.7, 64.5, 95.3, and 117.3) (Szafranski et al., 2016) and in two newly reported patients (147.3 and 158.3) map within ∼22.9 kb genomic interval between L1PA2 and L1PA3 harboring five different Alu elements (Figure 1; Table 1; Supporting Information, Figure S3). Three of these breakpoints (pts 64.5, 95.3, and 147.3) map to the same AluSx (chr16:86,287,015-86,287,326), one (pt 158.3) maps to AluY (chr16:86,284,317-86,284,617), and one (pt 117.3) maps to AluSx1 (chr16:86,288,115-86,288,338). All those Alu elements are directly oriented with regard to each other and their partners at the other breakpoints. Thus, those deletions represent Alu/Alu-mediated genomic rearrangements (Song et al., 2018). In patient 28.7, breakpoint-containing regions were narrowed by aCGH to chr16:86,140,499 and chr16:86,285,499, but could not be sequenced (Stankiewicz et al., 2009). The GC content of the identified microhomologies around the deletion breakpoints was 48% (SD ± 16%), similar to 54% (SD ± 2%) average GC content for those three Alus (Figure 2b). We found that the locations of deletion breakpoints do not correlate with the presence of a particular Alu stem-loop structure. AluY and AluSx each harbor PolIII promoter regions, thus similarly as L1PA2 and L1PA3, they might be transcribed. None of seven copies of the 7-mer recombination-associated motif, located between L1PA2 and L1PA3, maps to Alu element.
Besides ACDMPV-causing deletions, query of the Database of Genomic Variants (DGV) database of polymorphic CNVs (https://dgv.tcag.ca/dgv/app/home) revealed 48 small, presumably nonpathogenic deletions, and three reciprocal duplications, all with breakpoints mapping within this ∼35 kb hotspot region (Figure 1). Although the breakpoints of those CNVs were not sequenced, based on aCGH data, the majority if not all of them are likely located within L1PA2, L1PA3, AluY, or AluSx.
Of note, the identified 16q24.1 instability hotspot resides in the intron 3 of an ∼61 kb-large lncRNA gene LINC01081 oriented in the same direction as all Alus and oppositely to L1s. All pathogenic CNV deletions discussed here arose de novo, on the maternally inherited chromosome 16. In one case (pt 179.3), the parental chromosome origin of de novo CNV deletion was not determined.
3.2 Evolutionary origin of ACDMPV-linked L1PA2, L1PA3, and Alu elements
BLAST analyses of ACDMPV-linked L1PA2 and L1PA3 at 16q24.1 revealed that they share 97% sequence identity. PCR and in silico phylogenetic analyses of these L1s indicated that they arose in the human–chimpanzee–gorilla lineage after its split from the orangutan lineage, most likely 7–12 million years ago (Supporting Information, Figure S4). We confirmed by PCR the presence of L1PA2 orthologs in the syntenic genomic regions of chimpanzee and gorilla and their absence in orangutan and macaque. However, we were able to amplify an ortholog of human L1PA3 only from chimpanzee, which suggests evolutionarily more recent arrival of this L1 at 16q24.1.
Sequence comparison of the nonhuman primate genomic regions syntenic with the human 16q24.1 instability hotspot (https://genome.ucsc.edu; Supporting Information, Figure S5) showed that the presence of AluSx1 and AluSx in this region dates around the time of the establishment of the Old World Monkey and the New World Monkey clades, respectively. The evolutionarily youngest AluY was found in this genomic location only in humans. Interestingly, analysis of the database of polymorphic CNVs (Figure 1) showed that this AluY element may be polymorphic in different world populations.
4 DISCUSSION
4.1 LINE/Alu hotspot at the FOXF1 locus on 16q24.1
We describe a novel ∼35 kb in size genomic instability hotspot on 16q24.1 that includes two L1s, L1PA2 and L1PA3, and five Alus located in between. L1PA2, L1PA3, and AluSx are evolutionarily young elements that harbor recurrent breakpoints of both recurrent and nonrecurrent CNV deletions. We propose that recurrent DNA breaks in the described genomic instability hotspot might have been repaired using DNA sequence homology or homeology in other directly oriented full-length or truncated L1 partner (NAHR) or (ii) microhomology in shorter homologous or nonhomologous sequences (i.e., MMBIR, MMEJ, or SSA), or by nonhomologous end joining (Carvalho & Lupski 2016; Song et al., 2018; Table 1). Analyses of the SPAST locus at 2p22.3 also implicated Alus in generation of recurrent DNA breaks leading to nonrecurrent CNVs (Boone et al., 2014).
4.2 L1 and Alu features that may predispose the genome to local instability
We found that the location of the deletion breakpoints along L1PA2 and L1PA3 in 16q24.1 correlates with the length of homology shared by flanking L1s. Breakpoints of CNVs with full-length L1 at their ends are located closer to the 5′ end of L1, whereas breakpoints of CNVs with L1 only at one of their two breakpoints are located closer to the 3′ end of L1.
Grechishnikova and Poptsova (2016) bioinformatically predicted potential of the evolutionarily young L1HS and L1PA1-L1PA8 elements and Alu repeats to adopt stem-loop structure. For instance, three conserved stem-loop clusters could form at L1's 5′UTR, two in the middle of the ORF2, two at the end of ORF2, one at the 3′UTR, and numerous less conserved palindromes along the entire L1 length. We did not find correlation between location of L1PA2 and L1PA3 breakpoints and stem-loop structures, or G-quadruplex structures (Sahakyan, Murat, Mayer, & Balasubramanian, 2017). However, we have identified GC skewing along the length of L1PAs and Alus, suggesting more frequent presence of their DNA in a single-stranded form (due to, e.g., their relatively more frequent replication or transcription) that may be easier to fold into non-B DNA structures predisposing to DNA breaks.
It has been suggested that the high frequency of LINE- or Alu-mediated CNVs may result from replication–transcription collisions (Carvalho & Lupski 2016; Hastings, Lupski, Rosenberg, & Ira, 2009; Szafranski et al., 2016). We propose that secondary structures of L1s and Alus might contribute to those events by slowing down or stopping progression of transcription or replication. Transcription, especially of the longer genes, results in prolonged chromatin opening and formation of R-loops, and may persist into the S phase of the cell cycle, thus increasing the chance of replication fork stalling followed by illegitimate template switching or fork collapse with broken DNA ends (Hastings et al., 2009). The genomic instability hotspot described here overlaps a long noncoding RNA gene, LINC01081, transcriptionally codirectional with Alus and L1's antisense promoter. Such genomic arrangement may lead not only to replication–transcription, but also transcription–transcription collisions. Similarly, late replication increases chances of its interference with transcription, leading to stalled RNA polymerase complexes, and increasing the likelihood of template switching or the occurrence of DNA breaks within non-B DNA regions.
Of additional interest is general enrichment of 16q24.1 in recombination-associated 7-mer motif, in particular the presence of several copies of this motif within the described instability hotspot (one within each of the L1PAs and seven between them), suggesting that in some cases CNV formation might involve generation of double-strand breaks (DSBs), potentially initiated by meiosis specific SPO11 (Myers et al., 2008). Another possible scenario might involve generation of two DSBs in the vicinity of L1 elements, followed by resection of the annealing of two heterologous repeats by single strand annealing mechanism.
5 CONCLUSIONS
We demonstrate that the 16q24.1 genomic instability hotspot, harboring evolutionarily young L1s and Alus, predisposes the genome to formation of same- and variably-sized CNV deletions via both homology- and nonhomology-based mechanisms. As the detection of transposons and other repetitive elements is often challenging, we predict that a systematic genome-wide search for CNV breakpoint clusters will reveal more L1 and Alu genomic instability hotspots.
From the evolutionary perspective, TEs had contributed to development of hundreds of thousands of novel regulatory elements in the primate lineage and reshaped the human transcriptional landscape (Jacques, Jeyakani, & Bourque, 2015). More recently, Trizzino et al. (2017) speculated that TEs, including L1s and Alus, are the primary source of novelty in primate gene regulation. L1s and Alus appeared at 16q24.1 location relatively recently during primate evolution, and substantial fraction of CNVs that they mediate are nonrecurrent. We hypothesize that formation of variably-sized CNVs catalyzed by recurrent DNA breaks within TEs in unstable genomic loci may have even facilitated evolution of environmental adaptation when compared to the same-sized CNVs occurring by NAHR.
ACKNOWLEDGMENTS
This work was supported by grants awarded by the US National Heart, Lung, and Blood Institute (NIH grant R01HL137203) to P.St., the National Organization for Rare Disorders (2014 and 2016 16001 NORD grants) to P.Sz., and the Polish National Science Center (2012/06/M/ST6/00438) to A.G.
We thank Drs. Christine R. Beck, Grzegorz Ira, and James R. Lupski for helpful discussion.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
DATA DEPOSITION
CNV deletions associated with ACDMPV were submitted to dbVar (https://ncbi.nlm.nih.gov/dbvar): dbvar - ticket #28045-259747.