Germline variation in the oxidative DNA repair genes NUDT1 and OGG1 is not associated with hereditary colorectal cancer or polyposis
Communicated by David E. Goldgar
Abstract
The causal association of NUDT1 (=MTH1) and OGG1 with hereditary colorectal cancer (CRC) remains unclear. Here, we sought to provide additional evidence for or against the causal contribution of NUDT1 and OGG1 mutations to hereditary CRC and/or polyposis. Mutational screening was performed using pooled DNA amplification and targeted next-generation sequencing in 529 families (441 uncharacterized MMR-proficient familial nonpolyposis CRC and 88 polyposis cases). Cosegregation, in silico analyses, in vitro functional assays, and case–control associations were carried out to characterize the identified variants. Five heterozygous carriers of novel (n = 1) or rare (n = 4) NUDT1 variants were identified. In vitro deleterious effects were demonstrated for c.143G>A p.G48E (catalytic activity and protein stability) and c.403G>T p.G135W (protein stability), although cosegregation data in the carrier families were inconclusive or nonsupportive. The frequency of missense, loss-of-function, and splice-site NUDT1 variants in our familial CRC cohort was similar to the one observed in cancer-free individuals, suggesting lack of association with CRC predisposition. No OGG1 pathogenic mutations were identified. Our results suggest that the contribution of NUDT1 and OGG1 germline mutations to hereditary CRC and to polyposis is inexistent or, at most, negligible. The inclusion of these genes in routine genetic testing is not recommended.
1 INTRODUCTION
Hereditary forms of colorectal cancer (CRC) are caused by germline mutations or epimutations in the mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS2, EPCAM, APC, MUTYH, SMAD4, BMPR1A, STK11 and PTEN (Valle, 2014). In the last years, novel genes involved in CRC predisposition have been identified, such as POLE, POLD1, NTHL1, MSH3, and GREM1, as well as novel candidates that require further validation for their inclusion in routine genetic testing (Valle, 2017).
Some genes that encode proteins involved in base excision repair (BER), such as MUTYH and NTHL1, when mutated, cause a recessive form of adenomatous polyposis (Nielsen, Henry, Infante, & Brand, 2012 [Updated 2015 Sep 24]; Weren et al., 2015). However, the evidence gathered so far for other oxidative DNA damage repair genes, such as NUDT1 (HGNC-approved symbol, also called MTH1) or OGG1, is still insufficient to include their study in the routine genetic diagnostics of familial CRC and/or polyposis (Dallosso et al., 2008; Farrington et al., 2005; Garre et al., 2011; Kim et al., 2004; Kinnersley et al., 2014; Morak, Massdorf, Sykora, Kerscher, & Holinski-Feder, 2011; Sieber et al., 2003; Weren et al., 2015).
The major function of NUDT1 is to sanitize the nucleotide pool from oxidized nucleotides such as 8-oxo-dGTP (Nakabeppu, 2001; Nakabeppu, Kajitani, Sakamoto, Yamaguchi, & Tsuchimoto, 2006; Sakumi et al., 1993). NUDT1 catalyzes the hydrolysis of 8-oxo-dGTP, 8-oxo-dATP, and 2-HO-dATP to their corresponding monophosphate forms (Fujikawa et al., 1999; Sakumi et al., 1993). This prevents the incorporation of 8-oxo-dG into DNA, which has been demonstrated to cause genetic instability (Einolf & Guengerich, 2001; Kamath-Loeb, Hizi, Kasai, & Loeb, 1997). On the other hand, OGG1 (8-oxoguanine-DNA glycosylase 1) initiates the DNA BER pathway of oxidatively modified mutagenic purine base lesions like 8-oxo-G (Klungland & Bjelland, 2007).
With the aim of clarifying the involvement of NUDT1 and OGG1 in hereditary CRC, here we evaluate the presence of germline mutations in those two genes in unexplained MMR-proficient nonpolyposis familial CRC and polyposis cases, and investigate the functional effect of the identified genetic variants at DNA, protein, and cellular levels.
2 MATERIAL AND METHODS
2.1 Patients
A total of 529 CRC families, including 441 uncharacterized MMR-proficient nonpolyposis families (60 Amsterdam-positive families) and 88 unrelated polyposis cases, were included in the study (Bellido et al., 2016). All families were assessed at the Hereditary Cancer Program of the Catalan Institute of Oncology, IDIBELL, and at the Familial Cancer Unit of the Spanish National Cancer Research Center (CNIO), between 1999 and 2012. In addition to the MMR status evaluation in the tumors for the nonpolyposis CRC cases, and the analysis of APC and MUTYH mutations in the unexplained polyposis patients, all included patients were screened for germline mutations in the proofreading domains of polymerases epsilon (POLE) and delta (POLD1) (Bellido et al., 2016), and for the recurrent c.268C>T (p.Gln90*) mutation in NTHL1 (Belhadj et al., 2017). Informed consent was obtained from all subjects and the study received the approval of the Ethics Committee of the Bellvitge Biomedical Research Institute (IDIBELL) (PR073/12).
2.2 DNA and RNA extractions
Genomic DNA from peripheral blood was extracted using the FlexiGene DNA kit (Qiagen, Hilden, Germany), and from formalin-fixed paraffin-embedded samples, using the QIAamp DNA FFPE tissue kit (Qiagen), following the manufacturer's instructions. RNA from cultured lymphocytes was extracted using a standard Trizol-based protocol.
2.3 Mutation identification in pooled samples
Patients were screened for OGG1 and NUDT1 mutations using a combination of PCR amplification in pooled DNAs and targeted massively parallel sequencing, as previously described (Bellido et al., 2017). Primers are shown in Supp. Table S1. Variants with a population minor allele frequency below 1% (source: GnomAD) were selected, hypothesizing either a dominant or recessive mode of inheritance.
2.4 Direct automated sequencing
Direct automated (Sanger) sequencing was used to validate the results obtained by massive parallel sequencing, identify the mutated cases within pools, and perform cosegregation studies and somatic mutation identification (Primer sequences in Supp. Tables S1 and S2). Sequencing was performed on an ABI Sequencer 3730 and data analyzed with SeqMan Pro v.13 (Lasergene, DNAStar, Madison, WI).
2.5 In silico predictions
The impact of the identified missense variants was analyzed by using the in silico algorithm CONDEL and 13 additional scores obtained from dbNSFP v3.5 (January 12, 2018), including SIFT, PolyPhen-2 (HVAR and HDIV), MutationTaster, MutationAssessor, PROVEAN, MetaSVM, MetaLR, M-CAP, REVEL, CADD, VEST3, and FATHMM-MKL (Kumar, Henikoff, & Ng, 2009; Liu, Wu, Li, & Boerwinkle, 2016). The effect on splicing was evaluated using Human Splice Finder v.3.0 (Desmet et al., 2009). Except for CONDEL, prediction data were provided by ANNOVAR (Wang, Li, & Hakonarson, 2010). A consensus interpretation of the in silico predictions was established according to the number of tools predicting a damaging or a neutral effect, requiring a minimum of 70% of the predictions, that is, 10 out of 14, agreeing in the variant classification. Otherwise, the impact of variant was considered as inconclusive.
The effect of the variants on the stability of the protein 3D structure was calculated using CUPSAT, I-Mutant 2.0, ERIS, PoPMuSiC, MAESTRO, and INPS-3D (Capriotti, Fariselli, Calabrese, & Casadio, 2005; Dehouck, Kwasigroch, Gilis, & Rooman, 2011; Gonnelli, Rooman, & Dehouck, 2012; Laimer, Hofer, Fritz, Wegenkittl, & Lackner, 2015; Parthiban, Gromiha, & Schomburg, 2006; Savojardo, Fariselli, Martelli, & Casadio, 2016; Yin, Ding, & Dokholyan, 2007).
2.6 Culture of lymphocytes and splicing analysis
The effect on splicing caused by NUDT1 c.-13+2T>A (NM_198950.1), NUDT1 c.143G>A, and OGG1 c.137G>A was evaluated in RNA extracted from peripheral blood lymphocytes (PBLs) of the corresponding carriers and controls cultured in presence and absence of puromycin. Genomic DNA was eliminated using DNAse I Amplification Grade kit (Invitrogen, Carlsbad, CA). Retrotranscription (RT) was performed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA). RT-PCR was carried out for the NUDT1 region comprised between exons 1–4 for c.-13+2T>A, and exons 1–3 for c.143G>A. OGG1 c.137G>A was analyzed by multiplex RT-PCR amplifying both alleles (exons1–3 for the wild-type [WT] allele and exon1–intron1 for the mutant allele). PCR products were run in a 1.5% agarose gel, visualized in a UV transilluminator and subsequently sequenced. Primers are detailed in Supp. Table S2.
2.7 NUDT1 allele-specific expression analysis
The region containing the NUDT1 c.-15T>A variant (transcript NM_198952.1) was PCR-amplified in genomic DNA and cDNA obtained from peripheral blood of a c.-15T>A carrier (transcript-specific primers). Allele-specific expression was assessed with the ABI PRISM SNaPshot Multiplex Kit (Applied Biosystems) (primers in Supp. Table S2). Allele-specific expression was measured using the height peak intensities and comparing the proportion of both alleles (T/A) between cDNA and gDNA (Valle et al., 2008).
2.8 In vitro assays to evaluate the functional impact of NUDT1 p.G48E and p.G135W
Detailed methodological procedures of the in vitro assays are described in Supp. Material. Briefly, NUDT1 mutant and WT constructs were produced and the corresponding proteins obtained. We evaluated the effects of p.G48E on NUDT1 activity (ability to hydrolyze dGTP) and on the thermal stability of the protein by differential scanning fluorimetry. Binding of NUDT1 inhibitors TH588 and TH1579 was assessed by inhibition and thermal stability experiments.
NUDT1 WT and p.G48E and p.G135W mutant constructs were obtained for mammalian expression for subsequent evaluation of their intracellular stability. Assessment of the proteasomal degradation of WT and mutant NUDT1 constructs was also performed.
2.9 Structural analysis
The structure of NUDT1-WT in complex with 8-oxo-dGMP (PDB: 3zr0) was previously analyzed (Svensson et al., 2011). The mutations p.G48E and p.G135W were introduced using Coot (Emsley & Cowtan, 2004; Emsley, Lohkamp, Scott, & Cowtan, 2010). All common rotamers of the side chains were analyzed for the most forgiving positioning. However, no rotamer could accommodate the side chains without severe clashes, therefore the most common rotamers were used. All structure figures were prepared using PyMOL (https://www.pymol.org).
2.10 Somatic 2nd hit analysis
Detailed description of the methods used for loss of heterozygosity (LOH) determination and promoter methylation analysis is shown in Supp. Material.
2.11 CRC case–control study
The frequency of OGG1 c.137G>A (p.R46Q) was assessed in a population-based multicase–control study (MCC-Spain, www.mccspain.org) (Castano-Vinyals et al., 2015). This study included 1,336 CRC patients and 2,744 cancer-free controls from Spain genotyped with the Illumina Infinium Human Exome Bead Chip array, which includes rare variants in coding regions observed in the 1000 Genomes project.
Fisher's exact test was used to evaluate genotype frequencies between cases and controls obtained in the MCC-Spain study.
3 RESULTS
3.1 NUDT1
One novel and four rare (population MAF < 1%) variants were identified in NUDT1 (NM_002452.3). Four were missense and one had a possible effect on splicing (Table 1). The phenotypes of the carrier families/probands include two nonpolyposis Amsterdam-positive families, two Bethesda and one classic adenomatous polyposis (>100 adenomas) (pedigrees in Figure 1).
| NUDT1 varianta | Protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NM_002452.3 (GRCh38) | NP_002443.3 | dbSNP (rs#) | Population MAF % (ESP6500/ExAC/GnomAD) | Evolutionary conservationb | Consensus score for functionalc and structurald predictions | Splicing prediction (HSF v3.0) | Second hit (NUDT1 status in tumor) | Functional studies | Cosegregation | Variant classificatione |
| c.7G>A | p.A3T | rs139296148 | 0.015/0.0045/0.0047 |
|
|
No change |
|
n.a. | Noncarrier: Sister (20 polyps, 53–57 y) | Likely benign |
| c.143G>A | p.G48E | n.a. | n.a. |
|
|
|
|
Variant affects MTH1 activity and stability | n.a. | Uncertain significance/Likely pathogenic |
| c.395A>T | p.K132I | rs146088002 | 0.15/0.076/0.075 |
|
|
No change | LOH | n.a. |
|
Uncertain significance/Likely benign |
| c.403G>T | p.G135W | rs182662727 | −/0.020/0.0177 |
|
|
No change |
|
Variant affects MTH1 stability |
|
Uncertain significance/Likely pathogenic |
| NC_000007.14:g.2242977T>A | rs1139425 | −/0.0052/0.002859 | n.a. | n.a. |
|
Variant affects splicing of NM_198950 creating expression imbalance of NM_198950 and NM_198952 | n.a. | Likely benign | ||
| c.-13+721T>A (NM_002452.3) | Intronic |
|
||||||||
| c.-15T>A (NM_198952.1) | 5′UTR |
|
||||||||
| c.-13+2T>A (NM_198950.1) | Splice-site |
|
- a NUDT1 variants have been submitted to the LOVD database (https://databases.lovd.nl/shared/variants/NUDT1/unique).
- b PhyloP score (values between −14 and +6): Sites predicted to be conserved are assigned positive scores. PhastCons score (values between 0 and 1): the closer the value is to 1, the more probable the nucleotide is conserved.
- c In silico functional prediction results are detailed in Supp. Table S3. Between brackets, number of predictions that support the indicated result (total number of predictions: 14).
- d 3D structure prediction: Protein stability calculations were performed using PoPMuSiCv3.1, ERIS, CUPSAT, I-Mutant 2.0, MAESTRO, and INPS-3D. Detailed results are shown in Supp. Table S4. Between brackets, number of predictions that support the indicated result.
- e According to the standard guidelines for the interpretation of sequence variants (recommendations of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology) (Richards et al., 2015).
- Abbreviations: CRC, colorectal cancer; D, damaging or deleterious; ESP6500, NHLBI GO Exome Sequencing Project; ExAC, Exome Aggregation Consortium; GnomAD, Genome Aggregation Database; HSF v3.0, Human Splicing Finder v.3.0; MAF, minor allele frequency; n.a., not available information.

NUDT1 c.7G>A (p.A3T) (rs139296148; MAFGnomAD = 0.0047%), present in an individual with CRC diagnosed at the age of 55 years and belonging to an Amsterdam-positive family, was not present in the proband's sister, who had undergone the removal of 20 polyps in her 50s. No cosegregation studies could be performed on the maternal branch of the family, where three additional CRC cases (age range: 40–60 years), among other tumors, had occurred (Figure 1, Family A). The variant, which affects a nonconserved amino acid, was predicted to be neutral by all in silico algorithms and to cause no splicing alterations (Table 1 and Supp. Table S3).
The novel variant, NUDT1 c.143G>A (p.G48E), was identified in an individual diagnosed with CRC at the age of 48 years who underwent the removal of one sessile polyp, one hyperplastic polyp and a tubular adenoma with low-grade dysplasia at the ages of 48, 39, and 55 years, respectively. The patient had no relevant familial cancer history, except for his father, diagnosed with prostate cancer at 88 (Figure 1, Family B). No somatic inactivation of the WT allele was detected in the proband's tumor. In silico prediction of the variant's effect resulted inconclussive according to the established criteria (eight out of 14 predictors classified it as deleterious) (Table 1 and Supp. Table S3). The substitution of p.G48 for a residue with a larger side chain causes loss of NUDT1 activity (Fujii, Shimokawa, Sekiguchi, & Nakabeppu, 1999). This suggests that a glutamic acid (E) in this position would have a similar effect. Furthermore, glycine (G) is unique in its ability to allow many protein main chain conformations, not possible for other amino acids. Structure analysis revealed that residue p.G48 is positioned close to the nudix box, the region responsible for magnesium binding and which holds the catalytic amino acids near to the protein surface. The p.G48E mutation introduces negative charge into a negatively charged area of the protein, likely to cause repulsion and destabilization. In line with this, in silico algorithms and protein modeling predict 3D structure destabilization caused by the amino acid change (Table 1; and Supp. Table S3, Supp. Table S4, and Supp. Figure S1).
NUDT1 c.395A>T (p.K132I) (rs146088002, MAFGnomAD = 0.075%) was identified in a patient diagnosed with CRC at the age of 43 years. LOH analysis in the proband's tumor revealed loss of the WT allele (Supp. Figure S2). In silico prediction algorithms provided inconclusive results and no splicing defects (Table 1 and Supp. Table S3). Although cancer, including a CRC and a gastric cancer, aggregated in the paternal branch, the NUDT1 variant was inherited from the mother, cancer-free at 73 years of age and without family history of CRC (Figure 1, Family C).
NUDT1 c.403G>T (p.G135W) (rs182662727; MAFGnomAD = 0.0177%) was identified in a patient diagnosed with CRC and two adenomas at the age of 42 years, and in his brother, who had had two adenomas with moderate-grade dysplasia removed at the ages of 42 and 47 years, and a bladder cancer diagnosed at 56. Their mother had been diagnosed with two synchronic CRCs at the age of 59 years and in their maternal family branch, four aunts and one cousin had also been diagnosed with CCR (age range: 61–82 years). Cosegregation analysis revealed that one of the proband's maternal aunts, diagnosed with CRC at the age of 75 years, and one of his cousins, diagnosed with CRC at the age of 61 years, were noncarriers (Figure 1, Family D). No additional somatic NUDT1 alterations were detected in the proband's tumor. The variant, affecting a highly conserved amino acid, was predicted to be deleterious by 12 out of 14 in silico algorithms (Table 1 and Supp. Table S3). In the protein, residue p.G135 is located in a hydrophobic region close to the part of the active site involved in recognition of the nucleotide base. Structure modeling revealed that p.G135W introduces large steric clashes and destruction of hydrophobic packing causing major destabilization (Supp. Figure S1).
The variant c.-13+721T>A (rs1139425; MAFGnomAD = 0.0052%), located in intron 1 of the main transcript (NM_002452.3), was identified in a CRC patient with classic adenomatous polyposis (mucosa completely covered with polyps) diagnosed at the age of 46 years and no mutations in APC, POLE, or POLD1 or biallelic mutations in MUTYH, NTHL1, or MSH3. No somatic NUDT1 alterations were detected in the proband's tumor. Due to sample unavailability, we could not assess the carrier status of the proband's mother and father, diagnosed with breast cancer at the age of 55 years and prostate cancer at the age of 54 years, respectively (Figure 1, Family E). The intronic NUDT1 c.-13+721 variant does not alter the splicing of the main transcript (NM_002452.3) (Figures 2a and 2b), however, it affects the splicing (c.-13+2T>A) of a shorter secondary transcript, NM_198950.1, causing the inclusion of intron 2 (Figure 2a). Transcript-specific cDNA amplification revealed that the inclusion of intron 2 results in the creation of a longer transcript identical in sequence and size to the longest NUDT1 transcript, NM_198952.1, thus causing a transcript imbalance (Figure 2c). This was further confirmed by assessing the allele-specific expression of c.-15T>A (NM_198952.1, longest transcript) in the carrier's lymphocytes (Figure 2d). Despite the increased expression of c.-15A (NM_198952.1) allele, the effect of this alteration, located in the upstream region of the transcript, remains unknown. Moreover, own RNA-seq data from normal colon mucosae obtained from 180 CRC patients showed that the expression of the affected transcripts, NM_198952.1 and NM_198950.1, is null or extremely low in normal colon tissue (Supp. Figure S3), further supporting the irrelevance of the variant in CRC predisposition. Leading to the same conclusion, c.-13+2T>C (NM_198950.1), affecting the same nucleotide as c.-13+2T>A, and therefore causing the same splicing alteration, has been reported at relatively high frequency in specific populations (e.g., Finnish or Asian MAFGnomAD = 3%), occurring even in homozygosity.
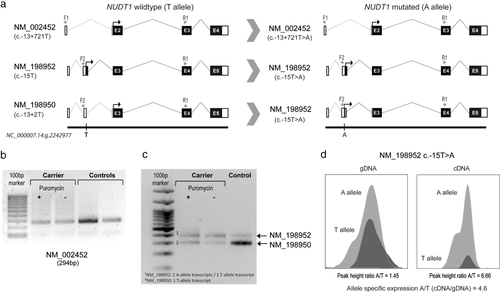
3.2 Effect of NUDT1 missense variants on protein activity and stability
Of the four NUDT1 missense variants identified, the two most highly conserved, c.403G>T (p.G135W) and c.143G>A (p.G48E), were the ones that most consistently showed prediction of altered function and/or structure destabilization. We performed several functional assays in order to validate these predictions and obtain more conclusive evidence of their potential pathogenicity.
In vitro assays to test the effect of the missense variant on NUDT1 activity and protein stability revealed that NUDT1-p.G48E displays a sixfold lower activity compared to WT when measured with dGTP (100 μM) as substrate (Figure 3a), and that the mutant protein is considerably less stable than NUDT1-WT with a melting temperature of 46°C compared with 56°C for the WT form (Figure 3b). In addition, the melting curve of NUDT1-p.G48E showed high relative fluorescence already at low temperature compared with NUDT1-WT, indicating that hydrophobic parts are exposed and suggesting that the NUDT1-p.G48E protein is not as tightly folded as the NUDT1-WT. Despite several attempts to produce NUDT1-p.G135W by using varying tags, expression strains and expression temperatures, we were not able to obtain NUDT1-p.G135W as a soluble protein, suggesting that this mutant is not folded correctly and is intrinsically unstable.

To better understand why the activity of NUDT1-p.G48E is lower than that of NUDT1-WT, we performed an in-depth kinetic analysis using the substrates dGTP and 8-oxo-dGTP. The results with 8-oxo-dGTP showed that p.G48E causes a drastic (14-fold) decrease on kcat, whereas Km (affinity for the substrate) remains similar (Figure 3c).
In order to investigate if the active site structure is altered in NUDT1-p.G48E compared with NUDT1-WT, we set out to determine the potency of the previously described NUDT1 inhibitors TH588 (Gad et al., 2014) and TH1579 (Warpman Berglund et al., 2016). IC50 values were determined from inhibition curves for NUDT1-WT and NUDT1-p.G48E with both inhibitors and were found not to be affected by the p.G48E mutation (Figure 4a). We also assessed the thermal stability of NUDT1-WT and NUDT1-p.G48E in presence and absence of TH588 and TH1579. NUDT1-WT and NUDT1-p.G48E are stabilized by 15°C and 21°C, respectively, in presence of 100 μM TH588 and TH1579 (Figure 4b). These data indicate that the part of the active site that is involved in binding to TH588 and TH1579 is not affected by the p.G48E mutation.

The profound effect on catalytic efficiency caused by the p.G48E mutation is in agreement with what was published before (Fujii et al., 1999). However, more or less identical Km values between mutant and WT enzymes as well as similar IC50- values for the tested inhibitors, imply that either only a fraction of the NUDT1-p.G48E preparation is correctly folded and holds an intact active site, or that only the region carrying the catalytic residues, but not substrate or inhibitor binding, is affected causing the drastic decrease in the kcat value.
We assessed the stability of NUDT1-p.G48E and NUDT1-p.G135W in a cellular context where cellular expression of the fusion proteins GFP-P2A-V5-NUDT1-WT/p.G48E/p.G135W was induced by adding Doxycycline. Once produced, the fusion proteins are cleaved into TurboGFP and V5-NUDT1-WT/p.G48E/p.G135W, resulting in equal levels of the two parts of the fusion protein. Assessment of the levels of the NUDT1 protein relative to TurboGFP using Western blot revealed that NUDT1-p.G48E and NUDT1-p.G135W were reproducibly less stable than NUDT1-WT, with NUDT1-p.G135W being the least stable in the cell (Figure 5).
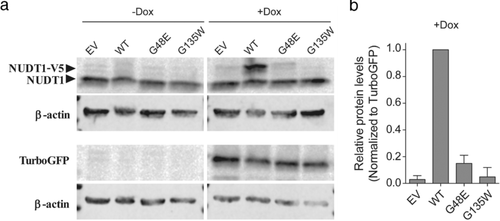
Since misfolded proteins are degraded by the 26S proteasome, we tested if that was the case for the NUDT1 proteins. Cells expressing NUDT1-p.G48E, NUDT1-p.G135W, and NUDT1-WT were treated with the proteasome inhibitor MG132, and protein levels were analyzed over time. We observed accumulation of NUDT1-WT and both mutants with time after inhibition, as well as accumulation of ubiquitin as a result of proteasome inhibition (Supp. Figure S4). Quantification of band intensity of NUDT1-WT and NUDT1-p.G48E before MG132 treatment and after 9 hr of treatment with MG132, showed that NUDT1-WT levels increased approximately threefold relative to GAPDH levels, whereas NUDT1-p.G48E increased about ninefold, indicating that p.G48E is more rapidly degraded by the proteasome compared with NUDT1-WT. Since no NUDT1-p.G135W was visible before MG132 treatment, no such comparison was possible. However, NUDT1-p.G135W levels clearly increased upon proteasome inhibition and the reason for no detectable NUDT1-p.G135W is likely due to fast degradation of the protein by the proteasome (Supp. Figure S4).
3.3 NUDT1 variants in hereditary CRC and cancer-free population
If NUDT1 were a CRC predisposing gene, one would expect a higher frequency of NUDT1 mutations in familial/early onset CRC cohorts, including ours, compared with the frequency in the general population. To evaluate the presence of germline NUDT1 variants in European population, we analyzed the publicly available data from 4,300 Caucasians (European American) analyzed for ESP6500SI-V2 (not related to cancer). We considered all individuals carrying nonsynonymous genetic variants (reference transcript NM_002452.3), that is, stop-gain, frameshift, and missense changes, as well as variants affecting canonical splice-sites, with MAF < 1%. Under these premises, 30 of the 4,300 individuals (0.70%) were carriers of predicted deleterious NUDT1 variants. Following the same criteria, in the familial CRC series herein studied (441 nonpolyposis CRC families and 88 adenomatous polyposis cases), we identified four (0.75%) carriers of nonsynonymous variants, being this frequency similar to the one observed in noncancer population. To validate these results, we evaluated the data obtained from sequencing 1,006 exomes from familial CRC cases, publicly available at https://canvar.icr.ac.uk. In this series, the frequency of NUDT1 (NM_002452.3) variants (selection criteria indicated above) was 0.1%, or 0.89% if filtered (nonpass) variants were included, in line with the findings obtained in our study, where no evidence for NUDT1 as a hereditary CRC genes was found after the study of 441 nonpolyposis CRC families and 88 adenomatous polyposis cases.
3.4 OGG1
The only variant identified in the OGG1 gene (NM_016821.2) with a population MAF < 1% was c.137G>A (p.R46Q) (rs104893751; MAFGnomAD = 0.22% (600/275,590)). It had been previously shown that this variant might affect splicing causing the insertion of intron 1 (Garre et al., 2011; Kohno et al., 1998), having been associated with CRC predisposition when occurring in the germline (Garre et al., 2011).
In our cohort, c.137G>A (p.R46Q) was identified in 7 MMR-proficient CRC families; one fulfilling the Amsterdam criteria and the other six, the Bethesda guidelines (Supp. Table S5). Cosegregation analyses carried out in three carrier families identified four additional carriers of the OGG1 variant: three CRC-affected (ages at diagnosis: 47, 60, and 87 years) and a 54-year-old cancer-free individual. All in silico algorithms used predicted a deleterious effect for c.137G>A (p.R46Q) (Supp. Table S3), and the splicing predictor Human Splicing Finder v3.0, a change at the donor site 1 bp downstream (−11.2%) and the creation of a new acceptor site (+51.04%). In fact, as mentioned above, it had been previously shown that this variant affects splicing causing the insertion of intron 1 (Garre et al., 2011; Kohno et al., 1998). Due to the relatively high presence of c.137G>A among the MMR-proficient CRC families studied (observed MAF = 0.79% (7/882)) compared with the frequency reported in public databases (e.g., MAFGnomAD = 0.22% (600/275,590), we further studied this variant from both functional and population standpoints.
The insertion of intron 1 was analyzed in DNase-treated and nontreated RNA extracted from PBLs cultured in the presence and absence of puromycin of two carriers of OGG1 c.137G>A and three controls (noncarriers of c.137G>A). The transcript with the intron 1 insertion was not identified in the cases and controls treated with DNase, suggesting that the previously described alternative OGG1 transcript corresponded to genomic DNA contamination (Supp. Figure S5).
Informative LOH data were obtained from four tumors developed by unrelated carriers, revealing equal representation of both alleles.
Data from a CRC case–control study that included 1,348 CRC patients and 2,743 controls, all of them of Spanish (European) origin, showed that the OGG1 variant (rs104893751) was equally represented among cases and controls (Supp. Table S6). Moreover, among the studied cases there was a homozygous c.137G>A female carrier who had been diagnosed with CRC at the age of 75 years. Her father died at the age of 53 years from a gastric cancer and her mother from lung cancer at 81 years old. The phenotype of the homozygous carrier (CRC at the age of 75 years) does not reveal the aggressive phenotype one would have expected if the variant were pathogenic in heterozygosity.
4 DISCUSSION
In contrast to MUTYH and NTHL1, whose causal implication in CRC and polyposis predisposition is well documented, the evidence gathered so far for other genes implicated in oxidative DNA repair, such as OGG1 and NUDT1, is still inconclusive (Dallosso et al., 2008; Farrington et al., 2005; Garre et al., 2011; Kim et al., 2004; Kinnersley et al., 2014; Morak et al., 2011; Nielsen et al., 2012 [Updated 2015 Sep 24]; Sieber et al., 2003; Weren et al., 2015). In our cohort, comprised of 441 MMR-proficient nonpolyposis families and 88 unrelated polyposis cases, a total of five novel or rare (population MAF < 1%) variants in NUDT1 were identified, two of which, c.143G>A (p.G48E) and c.403G>T (p.G135W) were classified as potential pathogenic mutations based on strong experimental evidence of functional defect. The inconclusive or contradictory cosegregation data in the families led us to compare the prevalence of NUDT1 mutations in familial CRC, and control or population-based individuals. The similar frequency of nonsynonymous, loss-of-function and splice site NUDT1 genetic variants in cases and controls, suggests a lack of association of NUDT1 mutations with CRC predisposition.
By screening the NUDT1 gene in 81 patients with polyposis or hereditary nonpolyposis CRC, Morak et al. (2011) identified two unrelated individuals with the novel heterozygous NUDT1 c.295G>A (p.D99N) and c.346A>T (p.M116L) variants, although neither experimental evidence of pathogenicity nor cosegregation data were provided. A similar scenario was observed for NUDT1 c.92G>A (p.R31Q), which was identified in two unrelated patients in combination with MUTYH pathogenic mutations (Farrington et al., 2005). In contrast, other studies (all of them in small cohorts) did not identify potentially pathogenic NUDT1 germline variants (Dallosso et al., 2008; Garre et al., 2011; Kim et al., 2004; Sieber et al., 2003).
Even if two functionally relevant NUDT1 mutations were identified in our cohort, the weak cosegregation evidence, low frequency and similar presence of rare NUDT1 variants in cancer-free population, suggest that NUDT1 heterozygous (monoallelic) mutations do not cause a CRC and/or polyposis predisposing syndrome. The profound effect of the p.G48E mutation on NUDT1 activity and stability, and the pronounced effect of p.G135W on protein stability, suggest a major impact of these mutations on cellular sensitivity to oxidative stress in cells carrying these mutations. A defective NUDT1 function would lead to the accumulation of oxidatively damaged nucleotides (Fujikawa et al., 1999; Sakumi et al., 1993; Yoshimura et al., 2003), such as 8-oxo-G, which, if incorporated into DNA, would primarily cause G:C to T:A transversions (Yasui et al., 2014), eventually leading to carcinogenesis. However, the presence of the mutations in heterozygosity, together with the absence of somatic inactivation of the WT NUDT1 allele in the tumors, suggests that the presence of a fully functional copy of NUDT1 is sufficient to protect the cells from the deleterious effect of oxidized nucleotides. In this context, it could be anticipated that the biallelic inactivation of NUDT1 in the germline might cause a cancer-predisposing syndrome. However, the scarcity of heterozygous carriers of pathogenic mutations together with the absence of biallelic mutation carriers in large clinically selected cohorts (441 nonpolyposis CRC families and 88 polyposis patients (this study) and 1,006 familial CRC cases (https://canvar.icr.ac.uk), suggest that the existence of biallelic carriers in the population is extremely rare.
Regarding OGG1, only one novel/rare variant, c.137G>A (p.R46Q), was identified in our cohort. It was detected in seven unrelated MMR-proficient nonpolyposis CRC families. We experimentally demonstrated that this genetic change does not cause any splicing alterations, as had been previously suggested (Garre et al., 2011; Kohno et al., 1998). The case–control data presented in our study clearly indicate that c.137G>A (p.R46Q) is not associated with CRC. Moreover, a homozygous carrier (diagnosed with CRC at the age of 75 years), identified through the case–control study, did not show the aggressive phenotype expected for a mutation that is already pathogenic in heterozygosity. Our data suggest that the contribution of germline mutations in OGG1 to hereditary CRC and to polyposis cases is inexistent or, at most, negligible.
Limitations to this study include: (a) the limited sample size (441 nonpolyposis CRC families and 88 adenomatous polyposis cases) makes the observed findings plausible but not definitive; (b) the analysis of only MMR-proficient hereditary nonpolyposis CRC, which, although improbable due to the type of mutations induced by deficient oxidative DNA repair, might have missed NUDT1- or OGG1-mutated cases among MMR-deficient CRC patients; and (c) in the course of the study some variants were dismissed based on the lack of segregation with cancer in the carrier families; however, they might still be exerting an effect in combination with other variants in other cancer predisposing genes (di-/oligo-genic effect).
In conclusion, the scarcity or absence of germline pathogenic mutations in NUDT1 and OGG1 in yet-to-be-characterized hereditary MMR-proficient nonpolyposis CRC and/or polyposis cases, together with the similar frequency of rare variants in cancer-free controls and familial cases, suggests a lack of association of NUDT1 and OGG1 mutations with hereditary CRC and/or polyposis. Our findings, pending confirmation in additional series, do not support the analysis of NUDT1 and OGG1 for routine genetic diagnostics in hereditary CRC and/or polyposis.
ACKNOWLEDGMENTS
We thank the participating patients and families, the Units of Genetic Counseling and Genetic Diagnostics of the Hereditary Cancer Program of the Catalan Institute of Oncology (IDIBELL), Kristina Edfeldt and Camilla Sjögren for administrative assistance, and Sabina Eriksson and Flor Pineiro for technical assistance. We thank Tobias Koolmeister and Martin Scobie for the synthesis of the NUDT1 inhibitors TH588 and TH1579.
DISCLOSURE STATEMENT
The authors declare no conflict of interest.