Performance of in silico tools for the evaluation of p16INK4a (CDKN2A) variants in CAGI
Contract grant sponsor: European Cooperation in Science and Technology (COST Action BM1405) (NGP-net); Associazione Italiana per la Ricerca sul Cancro (AIRC) (grants MFAG12740, IG17753); Italian Ministry of Health (Ministero della Salute, grants GR-2011-02347754, GR-2011-02346845); National Health and Medical Research Council of Australia (1059775, 1083450); NIH (U41 HG007346, R13 HG006650).
For the CAGI Special Issue
Abstract
Correct phenotypic interpretation of variants of unknown significance for cancer-associated genes is a diagnostic challenge as genetic screenings gain in popularity in the next-generation sequencing era. The Critical Assessment of Genome Interpretation (CAGI) experiment aims to test and define the state of the art of genotype–phenotype interpretation. Here, we present the assessment of the CAGI p16INK4a challenge. Participants were asked to predict the effect on cellular proliferation of 10 variants for the p16INK4a tumor suppressor, a cyclin-dependent kinase inhibitor encoded by the CDKN2A gene. Twenty-two pathogenicity predictors were assessed with a variety of accuracy measures for reliability in a medical context. Different assessment measures were combined in an overall ranking to provide more robust results. The R scripts used for assessment are publicly available from a GitHub repository for future use in similar assessment exercises. Despite a limited test-set size, our findings show a variety of results, with some methods performing significantly better. Methods combining different strategies frequently outperform simpler approaches. The best predictor, Yang&Zhou lab, uses a machine learning method combining an empirical energy function measuring protein stability with an evolutionary conservation term. The p16INK4a challenge highlights how subtle structural effects can neutralize otherwise deleterious variants.
1 INTRODUCTION
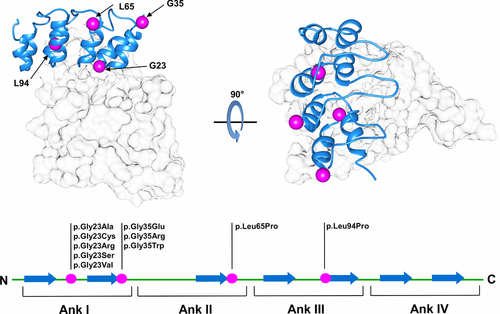
As genetic tests become routinely applied to the investigation of disease-associated variants, relevant efforts are made by the scientific community to develop computational tools for genetic variant evaluation (Niroula & Vihinen, 2016). A number of methods presenting different strategies have been presented, and their application is becoming a common routine in cancer research (Kannengiesser et al., 2009; Miller et al., 2011). In silico predictors are generally designed to provide a fast simplified response when compared with experimental screening protocols. However, lack of properly validated benchmarking represents the main limiting factor hampering wider application in a clinical scenario (Walsh, Pollastri, & Tosatto, 2016). Variants affecting tumor-suppressor genes, such as TP53 (Liu & Bodmer, 2006), VHL (Leonardi, Martella, Tosatto, & Murgia, 2011), and CDKN2A (Scaini et al., 2014) are actively investigated and collected in freely accessible databases (Forbes et al., 2015; Tabaro et al., 2016; Wang et al., 2015). However, the correct interpretation of their pathogenic significance is far from definitively addressed. One relevant issue remains our ability to correctly predict disease-causing gene variants among variants of unknown significance (VUS) (Wang & Shen, 2014). Correct prediction of susceptibility variants can foster the identification of molecular pathways causative of human diseases, particularly when variants affect well-understood genes previously validated by functional studies (Manolio, 2010). Since 2010, the Critical Assessment of Genome Interpretation (CAGI) experiment tries to objectively assess the state of the art of computational tools developed for genotype–phenotype determination. Here, we present a critical assessment of pathogenicity predictors applied to variants from the CDKN2A (MIM# 600160) tumor suppressor, also known as p16. CDKN2A is the major susceptibility gene identified in familial malignant melanoma. Approximately 40% of melanoma-prone families worldwide have CDKN2A germline variants (Hussussian et al., 1994). The CDKN2A locus maps to chromosome 9p21 and its regulation is particularly complex, involving alternative promoters, splicing, and reading frames of shared coding regions. Two structurally unrelated tumor suppressors, p16INK4a and p14ARF, involved in cell cycle regulation, are coded by alternative splicing of different first exons (1-α and 1-β). p16INK4a is a cyclin-dependent kinase (CDK4/6) inhibitor and p14ARF acts in TP53 stabilization, binding, and sequestering the MDM2 proto-oncogene (Serrano, Hannon, & Beach, 1993; Zhang, Xiong, & Yarbrough, 1998). Thus, alterations of this single locus compromises two important tumor-suppressor pathways at the same time (Andreotti et al., 2016; Aoude, Wadt, Pritchard, & Hayward, 2015). When associated with D-type cyclins, CDK4/6 promotes cell cycle progression through the G1 phase by contributing to the phosphorylation and functional inactivation of retinoblastoma-associated protein (Sherr, 1994; Weinberg, 1995). Structurally, p16INK4a consists of four repeated ankyrin-type motifs, composed of two antiparallel helices and a loop forming the CDK4/6-binding interface (Fig. 1). In the context of pathogenicity prediction, the ankyrin fold is challenging. Ankyrin repeats stack against one another to form a unique elongated single domain, with a multistate folding pathway conferring high structural plasticity. This highly modular nature confers unique characteristics such as a high affinity for protein–protein interactions (Tang, Guralnick, Wang, Fersht, & Itzhaki, 1999). However, stack modularity can also be seen as a gradient of transiently folded states, where a single amino acid substitution may be able to interrupt p16INK4a-specific periodicity, causing a severe perturbation of the entire protein structure (Peng, 2004). For this CAGI challenge, participants were asked to predict the effect of 10 CDKN2A variants in the p16-challenge, previously validated in cell proliferation rate assays. Twenty-two predictions using different strategies, for example, scoring functions based on sequence conservation, or machine learning predictors, were assessed. The results allow us to propose where pathogenicity prediction might be improved, as methods combining information from different strategies were found to have the most promising results.
2 METHODS
2.1 Dataset and classifications
The challenge includes 10 nucleotide variants affecting only the CDKN2A gene-coding region without interfering with p14ARF. Each variant codes for a single amino acid substitution, with no insertions or deletions. The variant nomenclature used in this work refers to CDKN2A mRNA isoform1 (GenBank identifier: NM_000077.4). Participants were requested to perform predictions of the cellular proliferation rate for each of the 10 mutant proteins as a percentage of the proliferation rate relative to pathogenic mutants (Table 1). A proliferation rate of 100% is used for pathogenic variants (positive controls), and 50% for wild-type-like variants (negative controls). Predictors were also allowed to specify a prediction confidence (standard deviation) for each variant, with a maximum of six alternative submissions per group. The standard deviation was only reported for 14 submissions, and the same confidence value was used for all predictions in five submissions. In a few cases, predictions have been manually rescaled during assessment as proliferation levels were wrongly reported as a fraction of 1 rather than 100 (where 100 represents the 100% positive control proliferation rate). A training set composed of 19 CDKN2A variants from Kannengiesser et al. (2009) and Miller et al. (2011) was also provided to the participants for training (Supp. Table S1). This choice was justified based on the similar use of bioinformatics tools to predict CDKN2A variant effects on cell proliferation as verified by experimental assays. Bioinformatics predictions were described to be comparable with verified real values for most variants (Kannengiesser et al., 2009; Miller et al., 2011). Real proliferation levels obtained from the literature were rescaled between 0.5 and 1 (proliferation level of wild-type and disease-like phenotypes, respectively).
| Proliferation rate | |||
|---|---|---|---|
| Nucleotide variant | Protein variant | Average | Standard deviation |
| c.67G>A | p.Gly23Ser | 0.69 | 0.04 |
| c.67G>C | p.Gly23Arg | 0.91 | 0.14 |
| c.67G>T | p.Gly23Cys | 0.86 | 0.13 |
| c.68G>C | p.Gly23Ala | 0.53 | 0.09 |
| c.68G>T | p.Gly23Val | 0.90 | 0.1 |
| c.103G>A; c.103G>C | p.Gly35Arg | 0.53 | 0.02 |
| c.103G>T | p.Gly35Trp | 0.86 | 0.09 |
| c.104G>A | p.Gly35Glu | 0.60 | 0.11 |
| c.194T>C | p.Leu65Pro | 0.66 | 0.1 |
| c.281T>C | p.Leu94Pro | 0.93 | 0.13 |
- Identifiers of variants affecting cell proliferation and relative proliferation level. Variant nomenclature refers to CDKN2A mRNA isoform1 (GenBank identifier: NM_000077.4); nucleotide numbering starts with the A of the ATG translation initiation site. Proliferation levels were rescaled between 0.5 (wild-type-like phenotypes) and 1 (tumor-like phenotypes).
2.2 In vitro proliferation assay of CDKN2A variants and data normalization
The experimental validation of the pathogenic effect of the variants used in CAGI is described in detail in Scaini et al. (2014). Briefly, the full-length CDKN2A cDNA was cloned in the pcDNATM3.1 D/V5-His-TOPO®_expression vector (Invitrogen, Life Technologies Corporation, Carlsbad, CA), engineered by site-specific mutagenesis (QuikChange® II XL Site-Directed Mutagenesis Kit; Stratagene, CA), and finally transfected in U2-OS human osteosarcoma cells (p16INK4a and ARF null, p53 and pRb wild type), as previously described (Scaini et al., 2009; Scaini et al., 2014). Three controls, no vector (G418 selection control), pcDNA3.1–EGFP (positive, variant-like control), and pcDNA3.1–p16INK4a wild type (negative control), were included in each experiment. All variants were independently tested at least three times. The proliferation rate was calculated as a percentage of the proliferation of variant-transfected cells (average of all replicates) at day 8 relative to the proliferation of EGFP-transfected cells, which was set as 100%. Transfection with wild-type CDKN2A induced a detectable, substantial growth inhibition (proliferation rate 50%), whereas various p16INK4a variants had different effects on cell proliferation, from wild-type-like to loss-of-function. The proliferation rates used for CAGI are shown in Table 1.
2.3 Performance assessment
Evaluating the performance of bioinformatics tools in predicting VUS impact is a non-trivial task. The assessment should not be seen as a mere discrimination of winners/losers, but rather aim at identifying which tool generated the most reliable prediction. A considerable number of performance measures were considered in order to perform a thorough assessment. The final goal was to generate a global overview of the strengths and weaknesses of each method. Correlation indices were considered first, as predictions are in a continuous range (cell proliferation rate). Both the Pearson correlation coefficient (PCC) and Kendall's Tau correlation coefficient (KCC) were calculated. Both range from +1 (perfect positive correlation) to −1 (perfect inverse correlation) with 0 representing a random performance. Root mean square error (RMSE) was calculated to better estimate the difference between predicted and real values. To further assess the prediction reliability in a medical setting, a binary classification was used. Proliferation levels were divided in two classes, benign and pathogenic, with three different proliferation thresholds suggested by the data provider, that is, potentially pathogenic (>65%), probably pathogenic (>75%), and likely pathogenic (>90%). The area under the ROC curve (AUC) for each classification threshold was also calculated. The standard deviation of the predicted proliferation rate was used to calculate the fraction of predictions within standard deviation (PWSD). To address the issue related to missing and very large confidence range, PWSD was calculated assuming a standard deviation of 10% for all submissions (PSWD10). The performance indices used in ranking are shown in Table 3, and additional performance measures at different thresholds can be found in Supp. Table S3. An overall ranking of predictors’ performance was defined as average ranking of four quality measures. All measures are defined in more detail in the Supp. Material. To assess the statistical significance of each performance index, 10,000 random predictions were generated and used to calculate an empirical continuous probability (score s), with a P value defining the proportion of random predictions scoring > s. The R scripts used to perform the assessment are publicly available from the GitHub repository at URL: https://github.com/BioComputingUP/CAGI-p16-assessment.
3 RESULTS
3.1 Participation and similarity between predictions
In the p16INK4a CAGI challenge, participants were requested to predict the effects of 10 p16INK4a VUS potentially causing malignant proliferation validated with cellular proliferation assays (Scaini et al., 2014). This challenge attracted 22 submissions from 10 participating groups, which were assessed without knowing the identity of the predictors. After the assessment was completed, only one group remained anonymous. Table 2 lists the participating groups, their submission IDs, and main features used for prediction. The majority of methods used evolutionary information derived from multiple-sequence alignments for prediction. Several methods also used the available crystal structure of p16INK4a bound to CDK6 (see Fig. 1) to calculate folding energies. Combinations of both approaches or of different predictors were also submitted. A summary for each method is described in the Supp. Material. Of the 10 participating groups, four contributed one prediction, one submitted two, four submitted three, and only one group submitted four different submissions.
| Submission ID | Group ID | Prediction features |
|---|---|---|
| Submission 1 | Anonymous | / |
| Submission 2 | Bromberg lab | Conservation, annotation |
| Submission 3 | Casadio lab | Conservation, gene ontology |
| Submission 4 | Lichtarge lab | Conservation |
| Submission 5 | BioFolD lab | Conservation, gene ontology |
| Submission 6 | Vihinen lab | Metapredictor |
| Submission 7 | Dunbrack lab | Protein structure |
| Submission 8 | Gough lab | Conservation |
| Submission 9 | Moult lab | Metaprediction |
| Submission 10 | Yang&Zhou lab | Conservation, folding energy |
| Submission 11 | Bromberg lab | Conservation, annotation |
| Submission 12 | BioFolD lab | Conservation, gene ontology |
| Submission 13 | Vihinen lab | Conservation, amino acid features, gene ontology |
| Submission 14 | Gough lab | Conservation |
| Submission 15 | Moult lab | Metaprediction |
| Submission 16 | Yang&Zhou lab | Conservation |
| Submission 17 | Bromberg lab | Conservation, annotation |
| Submission 18 | BioFolD lab | Metaprediction |
| Submission 19 | Gough lab | Conservation |
| Submission 20 | Moult lab | Metaprediction |
| Submission 21 | Yang&Zhou lab | Folding energy |
| Submission 22 | Yang&Zhou lab | Folding energy |
- For each submission, predictor and a summary of features used for prediction are indicated.
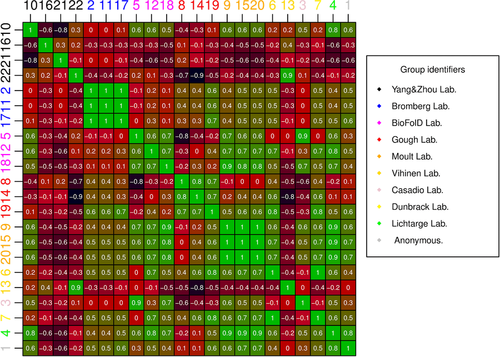
An analysis of prediction similarity was performed to better highlight the peculiarity of each submission (see Suppl. Fig. S1 for the full dataset). Almost all groups performing multiple submissions made very similar predictions (see Fig. 2). This is particularly evident for the Bromberg group, which were de facto mostly identical for many variants. A similar situation can be drawn for the Moult group, where a different fitting of two linear models (submissions 9, 15) produced identical predictions for most variants. A different rescaling process of submission 15 defined the third prediction (submission 20). Submissions 9 and 15 both predicted a majority of variants between 0.88 and 1. Predictions from the Gough and BioFolD groups are also quite strongly correlated among each other. Interestingly, submissions 5 and 3 (BioFolD and Casadio lab, respectively) are also highly correlated as both are based on two versions of the SNPs&GO method (Calabrese, Capriotti, Fariselli, Martelli, & Casadio, 2009; Capriotti et al., 2013). The Vihinen lab (submissions 6, 13) presents a weak anticorrelation among its predictions, probably due to predictions for all except one variant being very high (≥0.85). The four submissions from Yang&Zhou lab (10, 16, 21, 22) present almost no correlation, possibly also due to a sign error affecting three submissions.
3.2 Assessment criteria and performance measures
The type of insights to be gained from assessing a CAGI challenge depends strongly on the criteria used for evaluation. As this is a relatively novel field, extra care was given to this point. Ideally, the criteria should reflect the true performance of the methods, highlighting submissions that are of practical relevance. The simplest measures, binary classification and derived measures such as AUC, suffer from the choice of an arbitrary threshold, which may obfuscate interesting results. Correlation measures are good to indicate overall trends, but of little use to guide the selection of pathogenic cases as no threshold is used. At the other numerical extreme, RMSE is very clear, but can result in poor performance for all submissions. For an inherently continuous prediction challenge such as p16, determining the number of predictions within a fixed distance can arguably provide a measure combining features of binary classification and correlation. In order to understand better how related the assessment criteria are among each other, their correlation was plotted (Fig. 3). The PCC and KCC correlation coefficients are highly correlated with each other and with the three AUC measures. RMSE and two PWSD variants are less correlated and offer two alternative views of the data.
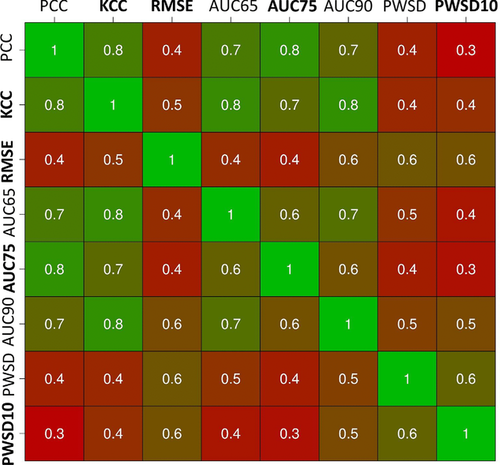
Using a reduced set of measures for the final ranking is suggested by the high pairwise correlation coefficients, suggesting they are measuring very similar features (see Fig. 3). A ranking including largely orthogonal measures should prove more robust and informative. For this reason, only four measures (one for each group) with low pairwise correlation were considered for the final ranking, that is, KCC, RMSE, AUC considering a 75% of proliferation threshold (AUC75), and PWSD considering a standard deviation of 10% for all submission (PWSD10). In particular, KCC was chosen as it is a rank-based measure appropriate when targets are continuous and their relative order is critical. The data provider recommended to use AUC75, as the corresponding proliferation level appeared to be the best threshold to separate pathogenic and neutral phenotypes. Finally, PSWD10 was preferred over PSWD as many predictors did not report standard deviation for their submissions.
3.3 Performance evaluation
The assessment of performance achieved by the 22 methods showed many predictions to have good results on average. This is particularly true considering AUC75, where most of the submissions achieved values between 0.7 and 1. For KCC, the average of the submissions shows a moderate to strong correlation with real data (see Table 3). Good results were however not sufficient for most predictions to be statistically significant. Very demanding thresholds emerged to separate significant results from random for this challenge, with only the top ranking methods being significant for most of the four performance indices (see below). This is probably due to the limited number of variants present in the test set, where wrong prediction of one variant corresponds to 10% of the dataset. Small variations in predictions could be reflected in remarkable fluctuation of performance indices due to the small number of variants considered. To perform a global assessment of predictor performance, we therefore decided to focus more on ranking than on numerical values achieved for each measure. Ranking variations not only may better reflect the magnitude of performance variation, but can also be considered more intuitive for nonspecialist readers. The Yang&Zhou lab (submission 10) performed best, ranking first in all performance indices except AUC75, where it is fifth (see Table 4). The Lichtarge lab (submission 4), an anonymous prediction (submission 1), and the Moult lab (submissions 15, 20) obtained higher AUC75 values. The Lichtarge lab also obtained good results considering KCC, where it ranked second. BioFolD (submission 5) also achieved good results, ranking second for both PSWD10 and RMSD and third for KCC. Furthermore, the BioFolD lab also performed well with submission 12, being second and third for PSWD10 and RMSD, respectively. Among the lower ranked predictions, an inverse correlation is found for submission 8 (−0.40), mainly resulting from low proliferation levels being predicted when real proliferation levels were high. Submissions 16 and 21 rank poorly, achieving an inverse KCC correlation (−0.56, −0.6). Notably, while all three submissions perform poorly, they probably followed opposed strategies. Submission 8 tends to be very conservative, with most of the predicted values close to a wild-type phenotype. Submissions 16 and 21 tend to be more biased toward the prediction of malignant phenotypes, with only one predicted value close to a milder phenotype. This trend seems to be shared among lower ranking predictions.
| Submission | PCC | KCC | RMSE | AUC65 | AUC75 | AUC90 | PWSD | PWSD10 |
|---|---|---|---|---|---|---|---|---|
| S1 | 0.83 | 0.45 | 23.51 | 0.81 | 1 | 0.76 | 5 | 3 |
| S2 | 0.33 | 0.02 | 21.29 | 0.57 | 0.62 | 0.55 | 3 | 2 |
| S3 | 0.53 | 0.47 | 25.5 | 0.83 | 0.7 | 0.64 | 2 | 2 |
| S4 | 0.84 | 0.63 | 16.48 | 0.81 | 1 | 1 | 4 | 5 |
| S5 | 0.66 | 0.6 | 15.81 | 0.9 | 0.88 | 0.9 | 7 | 6 |
| S6 | 0.23 | 0.34 | 25.67 | 0.57 | 0.58 | 0.79 | 2 | 3 |
| S7 | 0.22 | 0.2 | 18.2 | 0.57 | 0.68 | 0.62 | 3 | 4 |
| S8 | −0.34 | −0.4 | 39.21 | 0.19 | 0.42 | 0.26 | 1 | 1 |
| S9 | 0.7 | 0.38 | 20.18 | 0.86 | 0.88 | 0.71 | 3 | 3 |
| S10 | 0.83 | 0.69 | 9.24 | 1 | 0.92 | 1 | 7 | 7 |
| S11 | 0.33 | 0.02 | 21.29 | 0.57 | 0.62 | 0.55 | 2 | 2 |
| S12 | 0.57 | 0.47 | 15.93 | 0.67 | 0.84 | 0.9 | 4 | 6 |
| S13 | 0.11 | 0.05 | 20.08 | 0.57 | 0.42 | 0.64 | 5 | 5 |
| S14 | −0.22 | −0.4 | 23.29 | 0.19 | 0.42 | 0.26 | 5 | 5 |
| S15 | 0.76 | 0.51 | 18.83 | 0.86 | 0.96 | 0.81 | 4 | 3 |
| S16 | −0.45 | −0.56 | 22.48 | 0.12 | 0.08 | 0.14 | 2 | 2 |
| S17 | 0.43 | 0.25 | 21.8 | 0.67 | 0.72 | 0.57 | 2 | 2 |
| S18 | 0.46 | 0.28 | 16.35 | 0.67 | 0.72 | 0.76 | 6 | 2 |
| S19 | 0.3 | 0.07 | 20.3 | 0.45 | 0.76 | 0.55 | 2 | 3 |
| S20 | 0.76 | 0.51 | 17.7 | 0.86 | 0.96 | 0.81 | 4 | 4 |
| S21 | −0.62 | −0.6 | 23.71 | 0.19 | 0.12 | 0 | 2 | 2 |
| S22 | 0.15 | 0.2 | 18.45 | 0.6 | 0.4 | 0.76 | 3 | 3 |
- Results are shown for the main performance indices considered in the assessment.
- The top performing submission in each category is shown in bold and the second best is underlined.
| Rank | ||||||
|---|---|---|---|---|---|---|
| Submission | KCC | RMSE | AUC75 | PWSD10 | Average | Overall |
| S1 | 8 | 18 | 1 | 9 | 9 | 8 |
| S2 | 17 | 13 | 14 | 15 | 14.75 | 18 |
| S3 | 6 | 20 | 12 | 15 | 13.25 | 15 |
| S4 | 2 | 5 | 1 | 4 | 3 | 2 |
| S5 | 3 | 2 | 6 | 2 | 3.25 | 3 |
| S6 | 10 | 21 | 16 | 9 | 14 | 16 |
| S7 | 14 | 7 | 13 | 7 | 10.25 | 10 |
| S8 | 19 | 22 | 17 | 22 | 20 | 22 |
| S9 | 9 | 11 | 6 | 9 | 8.75 | 7 |
| S10 | 1 | 1 | 5 | 1 | 2 | 1 |
| S11 | 17 | 13 | 14 | 15 | 14.75 | 18 |
| S12 | 7 | 3 | 8 | 2 | 5 | 4 |
| S13 | 16 | 10 | 17 | 4 | 11.75 | 12 |
| S14 | 19 | 17 | 17 | 4 | 14.25 | 17 |
| S15 | 4 | 9 | 3 | 9 | 6.25 | 6 |
| S16 | 21 | 16 | 22 | 15 | 18.5 | 20 |
| S17 | 12 | 15 | 10 | 15 | 13 | 14 |
| S18 | 11 | 4 | 10 | 15 | 10 | 9 |
| S19 | 15 | 12 | 9 | 9 | 11.25 | 11 |
| S20 | 4 | 6 | 3 | 7 | 5 | 4 |
| S21 | 22 | 19 | 21 | 15 | 19.25 | 21 |
| S22 | 13 | 8 | 20 | 9 | 12.5 | 13 |
- Ranking of the different prediction methods based on performance indices in Table 1. To define the final ranking, average of ranking position for each performance index was used.
- The top performing submission in each category is shown as bold, whereas underlined is for the second best performance.
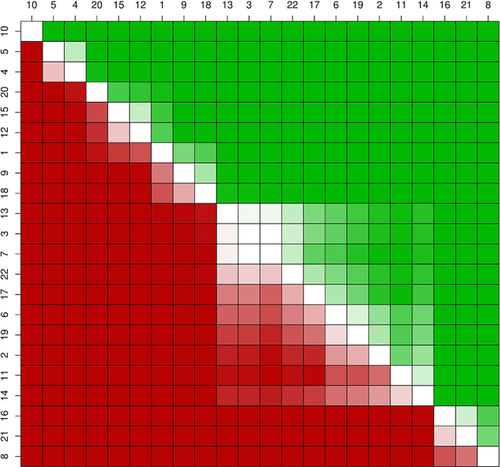
A statistical test of the average ranking over all four performance measures confirmed submission 10 (Yang&Zhou lab) as the best performer. No statistically significant difference can be identified between submissions 4 and 5 (Lichtarge, BioFolD; see Fig. 4) ranked second and third, respectively. A bootstrap simulation with 10,000 replicas was used to test whether the performance achieved by the three best submissions could be achieved by chance. Submission 10 performs better than random (P value < 0.05) for three out of four measures, the only exception being PSWD10. Submissions 4 and 5 perform better than random only considering KCC and AUC75 (see Table 5).
| S10 | S4 | S5 | |
|---|---|---|---|
| KCC | 0.015 | 0.015 | 0.015 |
| AUC75 | 0.029 | 0.004 | 0.048 |
| RMSE | 0.006 | 0.222 | 0.151 |
| PWSD10 | 0.059 | 0.389 | 0.183 |
- The P value for random predictions scoring better using each assessment metric is shown over 10,000 simulations.
- P values < 0.05 are shown as bold.
3.4 Difficult variants
An analysis of submissions shows prediction reliability to depend on position, with p.Gly23Ser, p.Gly35Glu, and p.Gly35Arg being particularly complex to address (see Supp. Table S2). p.Gly23Ser and p.Gly35Arg are the most mispredicted variants using PWSD10, with only two correct predictions. Both variants affect conserved positions that are known to have role in correct p16INK4a folding and CDK inhibition. A previous study (Scaini et al., 2014) addressing the same genetic changes showed p.Gly23Ser to introduce a weak interaction with S56. Although weak, this is thought to stabilize the overall fold, inducing a small local rearrangement of the p16-CDK4/6-binding interface. Predictions seem to miss this twofold effect. The p.Gly23Ser variant is mainly predicted as damaging, suggesting that current methods overpredict a pathogenic effect. A similar scenario can be seen for p.Gly35Glu and p.Gly35Arg. The G35 is a solvent-exposed residue, which localizes at the end of the first α-helix in the p16INK4a structure. Substitution of G35 with charged residues can be accommodated in the ankyrin fold, likely yielding neutral phenotypes (Scaini et al., 2014) mispredicted in this case. The only notable exception is submission 20, which shows the best accuracy with these difficult variants but misses most of the other variants. The p16INK4a challenge shows how different variants on the same residue can have widely diverging effects, which are not well predicted by many submissions.
4 DISCUSSION
Pathogenicity prediction of VUS is a challenging problem. It can manifest at different levels, such as protein function, subcellular localization, and pathways, as well as impairing multiple interactions a specific protein can exert with different partners (Hamp & Rost, 2012). Pathogenicity predictions are frequently performed through a priori knowledge of the biological problem, in most cases from an experimental characterization of disease-associated variants. In silico prediction can be considered a realistic benchmark of our understanding of these biological problems. Here, we presented results from the critical assessment of 22 different predictions in the CAGI p16INK4a challenge. Different submissions were compared to highlight the strengths and weaknesses of prediction strategies as applied to the human tumor-suppressor p16INK4a. The challenge had several peculiar characteristics. p16INK4a is a cancer-associated kinase inhibitor whose main function is protein–protein binding. It is also an ankyrin repeat protein, characterized by repetitive local short-range interactions (Peng, 2004; Scaini et al., 2014). In an ideal scenario, a reliable pathogenicity predictor should discriminate variations affecting both p16INK4a features. From a computational point of view, most predictors use position-specific scoring matrices (PSSM) and machine learning. The assessment suggests that our knowledge is sufficient to perform reliable predictions for the analyzed variants. However, relevant differences emerged among predictions. These differences stem in part from the strategy used for pathogenicity assessment. Others arise from expert knowledge, with similar approaches generating discordant predictions. Groups combining different strategies seem more robust when predicting CDKN2A variants. Predictions supplied from the Yang&Zhou lab are emblematic of this phenomenon. This group contributed four different submissions, rescaling PSSM value differences between wild type and variants, computing ΔΔG variation with ROSETTA3 (Dimaio, Leaver-Fay, Bradley, Baker, & André, 2011), computing ΔΔG with Dmutant (Zhou & Zhou, 2002) or combining them in a support vector machine using a linear kernel. Our assessment showed the Yang&Zhou lab reliability improving with prediction complexity (see Tables 3 and 4), peaking with the most complex submission 10. A similar reliability gradient was observed for other groups using different strategies, suggesting how a single method may be insufficient for pathogenicity prediction. Submission 10 presents the best fit with experimental data. On the other hand, a suboptimal AUC75 suggests the submission is less convenient for discriminating pathogenic from a wild-type-like phenotype. Conversely, submission 4 (Lichtarge group) presents the best AUC75 value, which may make it useful in a clinical setting. However, submission 4 predicts all variants as pathogenic at this threshold, which renders this method unreliable for clinical practice. Prediction performance seems to be also influenced by variant type. For example, variants affecting glycine 35 are on average easier to predict than glycine 23. The latter is known to be relevant for the correct ankyrin fold (Peng, 2004), as well as to localize at the p16INK4a/CDK4/6-binding interface (Miller et al., 2011; Scaini et al., 2014). For a generic pathogenicity predictor, this may be the worst case scenario. Sequence conservation analysis highlights the residue as conserved and relevant for protein structure, but may miss the pathogenic effect caused by interference at the protein–protein interaction interface. More advanced approaches, such as HMMs and neural networks, turned out to be the best strategies for this specific problem. It can be argued that the limited number of variants composing the dataset may limit generalization of the results and a larger set of variants might produce a different ranking. The dataset was chosen to represent a balanced ratio between pathogenic and neutral variants. Despite these intrinsic limitations, we believe this challenge may be representative of a clinical setting, where disease-associated genes are poorly described when it comes to variants found in patients. It is evident from the assessment that no method is able to perform errorless predictions. We expect the CAGI results to provide a starting point to improve the available methods and encourage using the scripts available on GitHub to help standardize the assessment.
ACKNOWLEDGMENTS
The authors are grateful to Francesco Tabaro for help with the assessment scripts.
Disclosure statement
The authors declare no conflicts of interest.




