Rare Variants in the Epithelial Cadherin Gene Underlying the Genetic Etiology of Nonsyndromic Cleft Lip with or without Cleft Palate
Contract grant sponsor(s): Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; CEPID Project 2013/08028-1); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 401952/2010-0); Fundos FEDER através do Programa Operacional Factores de Competitividade (COMPETE); Portuguese Foundation for Science and Technology (FCT; PTDC/SAU-ONC/110294/2009, PTDC/BIM-ONC/0171/2012, PEst-OE/EEI/LA0009/2013, and JF post-doctoral grant SFRH/BPD/87705/2012).
Communicated by Iain McIntosh
ABSTRACT
Nonsyndromic orofacial cleft (NSOFC) is a complex disease of still unclear genetic etiology. To investigate the contribution of rare epithelial cadherin (CDH1) gene variants to NSOFC, we target sequenced 221 probands. Candidate variants were evaluated via in vitro, in silico, or segregation analyses. Three probably pathogenic variants (c.760G>A [p.Asp254Asn], c.1023T>G [p.Tyr341*], and c.2351G>A [p.Arg784His]) segregated according to autosomal dominant inheritance in four nonsyndromic cleft lip with or without cleft palate (NSCL/P) families (Lod score: 5.8 at θ = 0; 47% penetrance). A fourth possibly pathogenic variant (c.387+5G>A) was also found, but further functional analyses are needed (overall prevalence of CDH1 candidate variants: 2%; 15.4% among familial cases). CDH1 mutational burden was higher among probands from familial cases when compared to that of controls (P = 0.002). We concluded that CDH1 contributes to NSCL/P with mainly rare, moderately penetrant variants, and CDH1 haploinsufficiency is the likely etiological mechanism.
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) and nonsyndromic cleft palate only (NSCPO) are two complex disorders within the nonsyndromic orofacial cleft (NSOFC) spectrum [Gorlin et al., 2001]. While the genetic etiology of NSCPO is largely unclear, genetic loci have been systematically implicated in NSCL/P, such as common low-risk 8q24, 10q25, and IRF6 variants [Rahimov et al., 2008; Birnbaum et al., 2009; Mangold et al., 2010, Brito et al., 2012aa, 2012b]. However, given the high heritability attributed to NSCL/P [Hu et al., 1982; Calzolari et al., 1988; Brito et al., 2011], searching for alternative genetic variants or mechanisms is necessary to bridge the missing heritability gap of these malformations.
Rare germline variants in the gene encoding the adhesion molecule epithelial cadherin, CDH1 (MIM# 192090), have long been associated with diffuse gastric cancer and lobular breast cancer [van Roy and Berx, 2008]. Most recently, CDH1 mutations have been reported in OFC patients in association with gastric cancer [Frebourg et al., 2006; Kluijt, et al. 2012; Benusiglio et al., 2013] or not [Vogelaar et al., 2013; Bureau et al., 2014]. These findings raise the questions as to what the proportion of NSOFC cases underlain by CDH1 variants and their attributed penetrance is, and which types of mutations or mechanisms lead to OFC, cancer, or both phenotypes.
Here, we performed a variant screening for CDH1 (NM_004360.3) coding region in 221 NSOFC probands (affected by NSCL/P [n = 189] or NSCPO [n = 32], either from nonfamilial [n = 138] or familial cases [n = 83]; Supp. Table S1). Sequencing was performed by using next-generation sequencing (NGS, exome or targeted gene sequencing) and Sanger sequencing (SS), and applied for 65 and 156 probands, respectively. Additional NGS or SS was performed for extra members of familial cases, when available. When exome sequencing was performed in affected members of the same family, we filtered out variants with minor allele frequency greater than 1% in public databases (1000 Genomes Project, and NHLBI ESP exomes) and in our in-house database of 609 Brazilian control exomes (Supp. Methods). Among the 221 probands, we identified a total of 47 variants, of which 12 were absent in our controls (two missense, one nonsense, and nine noncoding or synonymous variants; Supp. Table S2). Variants were submitted to the LOVD database (at http://www.lovd.nl/CDH1).
The novel missense variant c.760G>A (p.Asp254Asn, exon 6) was the most likely causative variant among the main candidates detected by exome analysis (mean coverage of 60×; average of 25,140 variants called for each individual; Supp. Table S3) in families F3788 and F617 (four affected individuals sequenced in each family; Supp. Fig. S1a and b). Both families segregate NSCL/P, and haplotype analysis of the exome data did not support a close relationship between these families (data not shown). SS of additional two affected and eight unaffected members from these two families supported segregation in accordance with an autosomal dominant model with incomplete penetrance estimated at 53%. Assuming this penetrance, a Lod score of 4.8 was obtained at recombination fraction (θ) 0, under an allele frequency of 0.0001. The other novel missense variant, c.2351G>A (p.Arg784His, exon 15), was found in the proband of family F1387 through SS. Segregation with NSCL/P was evidenced by its presence in three affected relatives (Supp. Fig. S1c), and penetrance was estimated at 62%. Further, the loss-of-function variant c.1023T>G (exon 8), predicted to create a stop codon at position 341 of CDH1 (p.Tyr341*), was found in the proband of family F7618 (Supp. Fig. S1d) through SS. Segregation with NSCL/P was suggested by its presence in an affected first cousin once removed. Although this represents the first association between this variant and NSCL/P, an association with hereditary diffuse gastric cancer (HDGC) has been previously observed [Guilford et al., 2010]. Penetrance was estimated at 31% in this family. Considering the four pedigrees, an overall 47% penetrance of NSCL/P was estimated, with a maximum Lod score of 5.86 at θ = 0 (individual Lod scores: F3788: 2.3; F617: 2.5; F1387: 0.9; F7618: 0.2).
Among the remaining nine noncoding or synonymous novel variants, only two variants were significantly scored by in silico tools for pathogenicity prediction (Supp. Table S4). Variant c.387+5G>A, which possibly decreases exon 3 splice donor site recognition, was found in a NSCL/P proband from a nonfamilial case (parental DNA unavailable for testing whether it is a de novo variant). We considered this variant as possibly pathogenic, although further functional studies are necessary. Variant c.2514C>T (exon 16), present in two unrelated probands (one isolated and one familial case), was discarded as pathogenic as it did not segregate with NSCL/P in the familial case.
E-cadherin consists of three major domains: a short cytoplasmic, a single transmembrane, and a large extracellular domain, with five repetitive subdomains [Paredes et al., 2012]. The p.Asp254Asn substitution is located at a calcium-binding site comprised by the amino acid sequence Asp–Gln–Asn–Asp, at position 254–257 of CDH1 [Tepass et al., 2000]. This site, in turn, is located in the outmost extracellular subdomain, which plays a major role in the molecular adhesive properties between cadherin trans-dimers [Shapiro et al., 1995]. Calcium binding in the extracellular subdomains is necessary for the cis-dimerization of E-cadherin, and for conferring rigidity to the extracellular domain [Nagar et al., 1996; Pertz et al., 1999]. Importantly, an amino acid substitution in a nearby calcium-binding site has been reported to completely suppress the cellular adhesive properties of E-cadherin in vitro [Ozawa et al., 1990]. The p.Arg784His substitution is located in the cytoplasmic domain, which is important for the assembly of catenins and for promoting cellular signaling [Nelson and Nusse, 2004]. Analyses with in silico tools indicated that these two missense variants are located in highly conserved regions of E-cadherin and probably impair protein function (Supp. Table S4). To determine their pathogenic potential in vitro, we transiently transfected Chinese Hamster Ovary (ATCC number: CCL-61) cells, which are negative for E-cadherin expression, with vectors encoding the wild-type (WT) E-cadherin and variants p.Asp254Asn and p.Arg784His (Supp. Methods). As revealed by Western blot and immunocytochemistry analysis, Asp254Asn cells showed decreased total E-cadherin protein expression (P = 0.00053; Fig. 1A), as well as reduced amount of E-cadherin located in the plasma membrane (Fig. 1B), when compared with cells expressing the WT protein. Furthermore, mutant cells were unable to form cellular aggregates and exhibited a scattered phenotype, contrary to the WT cells, thus clearly indicating impaired adhesive function (Fig. 1C). Even though no structural impact was predicted in the mutated CDH1 protein (performed with FoldX, http://foldx.crg.es/: ΔΔG = –0.81 kcal/mol), our in vitro assays suggest that p.Asp254Asn may lead to premature degradation, as shown for other cancer-related CDH1 pathogenic variants [Simoes-Correia et al., 2008, 2012; Figueiredo et al., 2013]. The functional effect of this variant could also be related to disturbances in calcium ion binding, given its location. Arg784His cells, in turn, showed no observable difference from WT cells in total E-cadherin amount, its location in the plasma membrane, and its adhesive behavior (Fig. 1A–C). However, this result should not be sufficient to rule out the pathogenicity of this variant, since this in vitro assay may not be able to detect other types of functional effects, such as changes in interactions with other proteins, altering subsequent signaling pathways.
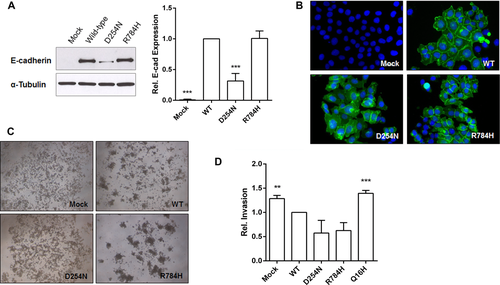
Families F3788 (p.Asp254Asn) and F1387 (p.Arg784His), the only two we were able to reascertain for cancer family history, include mutation carriers aged up to 70 years without cancer. This observation suggests that, under certain circumstances, CDH1 variants might cause NSCL/P alone. The invasive potential of Asp254Asn and Arg784His cells, investigated by an in vitro Matrigel matrix invasion assay, was similar to that of WT cells (Fig. 1D). Thus, it is possible that some E-cadherin mutations increase the risk of NSCL/P alone, while others increase risk of gastric cancer (mutations associated with higher invasiveness). However, it is of note that the current landscape of CDH1 mutations associated with gastric cancer and CL/P does not suggest any preferential distribution of mutations along the E-cadherin molecule (Fig. 2).

The overall prevalence of rare, possibly pathogenic CDH1 variants here reported was 2% (5 out of 221 NSOFC probands). To date, the CDH1 mutational repertoire in the literature associated with OFC includes 10 different mutations. Six of these mutations have been reported in families also segregating gastric cancer (four affecting mRNA splicing, one nonsense, and one frameshift deletion) [Frebourg et al., 2006; Kluijt et al., 2012; Benusiglio et al., 2013] and four were found in individuals with uncertain history of gastric cancer (one nonsense [Bureau et al., 2014] and three missense in a European cohort [Vogelaar et al., 2013]). Revisiting the list of variants described in the European cohort, we observed that the missense variant c.88C>A, which was reported in two patients from that study, was also found in two of our Brazilian controls; after removing this variant, the prevalence of possibly pathogenic CDH1 mutations in the European cohort becomes 2%, instead of the previously reported 5% and now similar to our estimate. Furthermore, considering that two of the possibly pathogenic mutations here reported (p.Asp254Asn and p.Tyr341*) and most of the 10 above-mentioned mutations are predicted to cause CDH1 loss-of-function, haploinsufficiency in critical stages of embryonic development seems to be the most likely mechanism by which rare variants in CDH1 lead to OFC.
To investigate whether the group of 221 NSOFC probands presents a higher burden of CDH1 rare variants compared to that of 609 Brazilian controls, we performed a gene-based sequence kernel association test (SKAT) [Wu et al., 2011]. A complementary two-tailed Fisher's exact test was performed to compare the proportion of individuals carrying at least one rare CDH1 variant between probands and controls. Only variants with minor allele frequency <1% and with nonneutral prediction in at least one in silico tool were selected for these tests. To avoid methodological bias in the tests, we only included variants from regions that were covered by both SS and NGS (with minimum coverage of 25×; Supp. Table S5). No significant differences in variant enrichment were detected by SKAT when comparing the 221 NSOFC probands with our 609 control exomes (P = 0.25). Similarly, no significant difference was detected in the number of individuals carrying these variants (two-tailed Fisher's exact test P = 0.85; patients: 9/221 [4%]; controls: 28/609 [5%]). Most of the probably pathogenic variants here reported (p.Asp254Asn, p.Tyr341*, and p.Arg784His) were found in familial cases with at least two affected members aside from the probands (4 out of 26 families matching the same condition, or 15.4%). Considering only probands from families with at least two additional affected individuals, significant differences were detected by SKAT (P = 0.002) and by two-tailed Fisher's exact test (P = 0.002; patients: 6/26 [23%]; controls: 28/609 [5%]). These findings suggest that the most noteworthy CDH1 etiological contribution to NSOFC arises from the fraction of NSCL/P cases involving moderate penetrance, which is best represented by familial cases. Since the previously suggested association between common variants and NSCL/P [Letra et al., 2009; Hozyasz et al., 2014] has not been supported by a large meta-analysis with GWAS data [Ludwig et al., 2012], rare variants seem to be the major contribution of CDH1 to NSCL/P etiology.
Given the lack of correlation between type/location of CDH1 rare pathogenic variants and NSCL/P or HDGC (Fig. 2), we speculate that a common underlying molecular mechanism could explain both phenotypes. Penetrance of CDH1 germline mutations implicated in HDGC depends on a second hit, which frequently occurs via promoter hypermethylation of the nonmutated allele, possibly triggered by environmental factors [Oliveira et al., 2009; Zeng et al., 2015]. In this regard, a lifetime exposure to such factors would be in agreement with the higher penetrance in HDGC (80%) [Pharoah et al., 2001], as compared to NSCL/P (47%). Germline, pathogenic variants in CDH1 could determine the resultant phenotype (NSCL/P or gastric cancer) under the influence of the following factors: time (early development or later in life), tissue (craniofacial or gastric structures), and exposure to environmental factors. In addition, given the prevalence of CDH1 pathogenic variants found in this study, we believe that the NSCL/P-associated CDH1 mutations are currently underrepresented, and future research should focus on their identification.
In summary, our results indicate a consistent role of rare, loss-of-function, moderately penetrant CDH1 variants in NSCL/P etiology. To better comprehend the mechanisms linking CDH1 to NSCL/P, as well as the risk of gastric cancer among NSCL/P individuals with mutations in CDH1, further studies are needed. Finally, CDH1 testing in NSCL/P familial cases should be discussed for genetic counseling purposes.
Acknowledgments
We thank the Operation Smile Brazil team and Dr. Daniela Bueno (Hospital Menino Jesus) for the productive collaboration, and Suzana Ezquina, Vitor Aguiar, Vanessa Simões, Dr. Paulo A. Otto, and Dr. Rui M. Ferreira for helpful discussions.
Disclosure statement
The authors declare no conflict of interest.




