Three Different Cone Opsin Gene Array Mutational Mechanisms with Genotype–Phenotype Correlation and Functional Investigation of Cone Opsin Variants
Contract grant sponsors: National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology; Fight for Sight (UK); Retinitis Pigmentosa Fighting Blindness (UK); Moorfields Eye Hospital Special Trustees; Foundation Fighting Blindness (USA).
Communicated by Nobuyoshi Shimizu
ABSTRACT
Mutations in the OPN1LW (L-) and OPN1MW (M-)cone opsin genes underlie a spectrum of cone photoreceptor defects from stationary loss of color vision to progressive retinal degeneration. Genotypes of 22 families with a range of cone disorders were grouped into three classes: deletions of the locus control region (LCR); missense mutation (p.Cys203Arg) in an L-/M-hybrid gene; and exon 3 single-nucleotide polymorphism (SNP) interchange haplotypes in an otherwise normal gene array. Moderate-to-high myopia was observed in all mutation categories. Individuals with LCR deletions or p.Cys203Arg mutations were more likely to have nystagmus and poor vision, with disease progression in some p.Cys203Arg patients. Three disease-associated exon 3 SNP haplotypes encoding LIAVA, LVAVA, or MIAVA were identified in our cohort. These patients were less likely to have nystagmus but more likely to show progression, with all patients over the age of 40 years having marked macular abnormalities. Previously, the haplotype LIAVA has been shown to result in exon 3 skipping. Here, we show that haplotypes LVAVA and MIAVA also result in aberrant splicing, with a residual low level of correctly spliced cone opsin. The OPN1LW/OPN1MW:c.532A>G SNP, common to all three disease-associated haplotypes, appears to be principally responsible for this mutational mechanism.
Introduction
Mutations in the long-(L-) (OPN1LW; MIM #300822) and medium-(M-) (OPN1MW; MIM #300821) wavelength-sensitive cone opsin genes on Xq28 [Nathans et al., 1986a, 1989; Neitz and Neitz, 2011] result in a number of cone photoreceptor disorders, including red-green color blindness (MIM #303900, MIM #303800), X-linked cone dysfunction (MIM #300843), X-linked cone dystrophy (COD5; MIM #303700), and blue cone monochromatism (BCM; MIM #303700) [Nathans et al., 1986a,b, 1989; Winderickx et al., 1992; Jagla et al., 2002; Crognale et al., 2004; Kellner et al., 2004; Neitz et al., 2004; Young et al., 2004; Michaelides et al., 2005a; Gardner et al., 2009; Gardner et al., 2010; Mizrahi-Meissonnier et al., 2010]. The majority of subjects with an L- and M-opsin disorder are thought to have a stable condition; however, some develop evidence of progression with increasing visual acuity loss, macular atrophy, retinal pigmentation, and electrophysiological evidence of S-cone and rod involvement. It thereby appears that some sequence alterations in the cone opsin genes can result in photoreceptor degeneration [Fleischman and O'Donnell, 1981; Nathans et al., 1989; Ayyagari et al., 1999; Michaelides et al., 2005b; Gardner et al., 2009; Gardner et al., 2010]. Recently, high-resolution retinal imaging in subjects with L- and M-opsin mutations suggested that phenotypic variation may be a result of subsets of mutations displaying distinct visual and cellular phenotypes [Carroll et al., 2009, 2010, 2012].
The L- and M-genes in a wild-type array lie in tandem, with a single L-gene upstream of one or more M-genes [Nathans et al., 1986a; Vollrath et al., 1988] with only the first two genes in the array thought to be expressed, owing to their proximity to an upstream cis-regulatory locus control region (LCR; MIM #300824) [Nathans et al., 1989; Wang et al., 1992]. The nucleotide composition of the first two genes within the array dictates the opsins expressed in L- and M-cone photoreceptors. The L- and M-opsin genes derive from a single opsin ancestor and retain a high-sequence homology of their coding sequences, introns, and downstream regions [Nathans, 1999]. The sequence similarity and their genetic head-to-tail arrangement results in frequent nonhomologous inter- and intragenic recombination (NHR) and gene conversion events [Hayashi et al., 1999]. As a result, whole genes are frequently deleted, gained, or rearranged to form hybrid L-/M- or M-/L-genes [Deeb et al., 1992; Neitz and Neitz, 2011]. Genetic rearrangement also forms combinations of polymorphic L- and M-variants that alter the spectral properties of the encoded proteins and lead to a range of normal or color vision deficient phenotypes [Neitz and Neitz, 2011].
BCM is a severe condition in which neither L- nor M-cones are fully functional. The genetic mechanism of BCM is thought to follow a number of pathways: one involves a single-step process in which an LCR deletion prevents expression of the genes in the array [Nathans et al., 1989; Wang et al., 1999; Smallwood et al., 2002]. Another involves a two-step process in which NHR reduces the number of cone opsin genes in the array to one, before a second event (mutation) inactivates the remaining gene [Nathans et al., 1993]. Reported inactivating mutations include missense and nonsense mutations and partial coding sequence deletions [Nathans et al., 1989; Ladekjaer-Mikkelsen et al., 1996; Wang et al., 1999; Ayyagari et al., 2000; Michaelides et al., 2005b; Gardner et al., 2009; Mizrahi-Meissonnier et al., 2010]. A further, less common mechanism involves inactivation of the first two genes of the array by a mutation that is transferred from one gene to another by means of NHR or gene conversion [Reyniers et al., 1995; Gardner et al., 2010]. More recently, an additional category of mutants has been identified that displays an unusual genetic mechanism. This category of ancestral interchange haplotypes has been identified as a significant cause of both red/green dichromacy and BCM [Carroll et al., 2004; Crognale et al., 2004; Neitz et al., 2004]. Interchange haplotypes arise as a result of recombinations between L- and M-opsin genes that create unusual, and potentially deleterious, combinations of exon 3 single-nucleotide polymorphisms (SNPs). In isolation, these SNPs would not be considered detrimental to gene function. One example is the deleterious SNP haplotype OPN1LW/OPN1MW: c.[ 457G; 511A; 513T; 521C; 532G; 538G] that encodes p.[Leu153; Ile171; Ala174: Val178; Ala180] or p.[L;I;A;V;A] for ease of reference.
Functional analyses of different cone opsin mutations suggest they may have different disease mechanisms and associated retinal phenotypes [Carroll et al., 2010; Gardner et al., 2010; Ueyama et al., 2012; Cideciyan et al., 2013]. Deletion of the LCR has been shown to result in a complete lack of expression of cone opsin, morphological changes to the cone outer segments, and to reduced cone density and disrupted organization of the cone mosaic [Carroll et al., 2010; Cideciyan et al., 2013]. In contrast, the missense mutations, p.Cys203Arg (p.C203R) and p.Trp177Arg (p.W177R), encode cone opsins that are misfolded and retained in the endoplasmic reticulum. These misfolded mutant cone opsins would be expected to be degraded leading to loss of cone opsin function and potentially might be toxic to photoreceptors, similar to misfolding mutations in rhodospin, leading to a potentially progressive cone dystrophy phenotype [Kazmi et al., 1997; Gardner et al., 2010; Carroll et al., 2012]. Interestingly, the disease-associated interchange SNP haplotype LIAVA has recently been shown to cause aberrant splicing in vitro, with skipping of exon 3. This mutational mechanism is predicted to result in a frameshift and a nonfunctional cone opsin [Ueyama et al., 2012].
In this study, we present detailed genetic and phenotypic analysis of a cohort of 26 affected males from 22 families with a range of phenotypes, from BCM to X-linked cone dystrophy, and describe correlations between the genetic mechanism of disease and clinical outcome.
Material and Methods
Subjects and Clinical Assessment
Male probands and participating relatives from 22 families with BCM, X-linked cone dysfunction syndrome, or X-linked cone dystrophy were recruited. If possible, individuals were examined more than twice over a period of 5 years or longer, allowing longitudinal evaluation of the phenotype. Written informed consent was obtained from all subjects and the study was conducted according to the tenets of the Declaration of Helsinki. Ethical approval for the study was provided by Moorfields Eye Hospital, London, the Women's and Children's Hospital, North Adelaide, and the Royal Victorian Eye and Ear Hospital, Melbourne.
Clinical assessment included medical history, family history, best-corrected Snellen visual acuity, refraction, slit-lamp biomicroscopy, dilated fundus examination, and digital fundus photography (TRC-501A; Topcon, Tokyo, Japan). Color vision was tested using Hardy–Rand–Rittler pseudo-isochromatic plates. Myopia was characterized as low (<3 diopters), moderate (3–6 diopters inclusive), or high (>6 diopters). Fundus autofluorescence (FAF) imaging and spectral domain optical coherence tomography (SD-OCT) imaging was performed where possible. The Spectralis HRA and OCT with viewing module version 5.1.2.0 (Heidelberg Engineering, Heidelberg, Germany) was used to obtain FAF and SD-OCT images and the HEYEX software interface (version 1.6.2.0; Heidelberg Engineering) was used to view images.
Electrophysiological assessment at Moorfields Eye Hospital included full field ERG and pattern ERG (PERG), recorded using gold foil corneal recording electrodes in 10 patients, incorporating the standards of the International Society for Clinical Electrophysiology of Vision (ISCEV) [Holder et al., 2007; Marmor et al., 2009; Bach et al., 2013]. A dark-adapted brighter flash ERG, as recommended by ISCEV, was additionally recorded to a flash strength of 11.0 cd s m−2 better to identify photoreceptor function. These patients also had scotopic red flash ERG testing, used to assess the relative contributions of dark-adapted cone and rod systems. Short-wavelength flash ERGs were performed using a blue stimulus (5 msec duration, 445 nm, 80 cd m−2) on an orange background (620 nm, 560 cd m−2) [Arden et al., 1999; Bach et al., 2013]. Full-field ERG measures of dysfunction were defined as: mild (70%–99% of normal amplitude), moderate (30%–69% of normal), severe (1%–29% of normal), or undetectable. Two subjects had repeat ERGs after 5 or 12 years to determine ERG progression. The ERGs in an additional nine children were performed with surface electrodes according to shortened pediatric protocols [Holder and Robson, 2006] that were sufficient to help establish the diagnosis.
Genomic Sequencing of the Cone Opsin Array
Genomic DNA was extracted from EDTA blood samples. The structural integrity of the L-/M-opsin gene array on Xq28 was investigated in several stages. Initially, all six coding exons and flanking intronic boundaries of OPN1LW and OPN1MW, and noncoding regions, including the 37-bp core sequence of the upstream LCR and the upstream and downstream promoters, were analyzed by direct sequencing of PCR amplicons, as described previously [Gardner et al., 2009]. The opsin cDNAs are numbered according to Ensembl Transcripts ENST00000369951 (OPN1LW) and ENST00000147380 (OPN1MW). cDNA nucleotide numbering uses +1 as the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1. PCR products were purified with ExoSAP-IT (GE Healthcare, Amersham, UK) and Sanger sequenced (Applied Biosystems, Warrington, UK) as described previously [Gardner et al., 2009]. Sequence data were examined with Lasergene DNA Star software (DNA STAR, Inc. Madison, WI). Sequence changes were tested for segregation with the condition in participating relatives.
Long-Range PCR to Determine Multigene Array Sequence and Structure
The exon structure of all intact genes including L-genes, M-genes, L-/M-, or M-/L-hybrid genes in each multigene array was determined by long-range PCR amplification of L-class genes (containing an L-type exon 5) and M-class genes (containing an M-type exon 5), followed by second round PCR amplification of exons 2, 3, and 4, using methods described previously [Neitz et al., 2004]. In cases where an unusual SNP haplotype was present in an L-class gene (comprising L-type exons 2–5), but not in any M-class genes, the position of L-class genes bearing the SNP haplotype was determined by testing whether the mutant L-gene was present in an upstream location, downstream location, or both. This was achieved by pairing a forward primer specific to the upstream promoter (UPF: CCTGGGCTTTCAAGAGAACCACATG) with an L-specific exon 2 reverse primer (LE2R; CCCAGCACGAAGTAGCCAG) and in a separate reaction, pairing LE2R with a forward primer upstream of OPN1LW and OPN1MW (LM1F; GGTGGGAGGAGGAGGTCTAA). The products were then sequenced to determine the presence of upstream and downstream sequence variants 5′ to exon 1. When an exon 3 mutant haplotype was present in a multigene array in more than one L-gene, and the array also had an M-gene with a normal exon 3 haplotype, it was concluded that in order for an L- and M-opsin deficient phenotype to occur, the L genes bearing the mutant haplotypes occupied the first two positions in the array.
LCR Deletions
In subjects in whom no amplification of the LCR-core sequence was achieved, a possible LCR deletion was suspected. Three sequence-tagged sites (STSs), located at intervals 7.5, 8.4, and 9.3 kb upstream of the L-gene transcription initiation site, were then amplified to determine the approximate upstream extent of the deletion, as described previously [Carroll et al., 2010]. If no product was obtained for the STSs, the first exons of the genes flanking the opsin array, MECP2 and TKTL, were amplified to evaluate the extent of the deletion (primer sequences available on request).
In Vitro Splicing Assay
Wild-type (color normal) or interchange OPN1LW (L-) and OPN1MW (M-) genomic sequence clones in pFLAGCMV-5a (minigenes), spanning exon 2–4, were described previously [Ueyama et al., 2012]. Five cone opsin minigenes were investigated in the current study: two with SNP haplotypes encoding color-normal opsins LVAIS or MVAIA, and three encoding the proposed deleterious haplotypes LIAVA, LVAVA, or MIAVA. The c.457C>A (p.Leu153Met) variant of the MIAVA haplotype was introduced into the L-opsin LIAVA minigene by site-directed mutagenesis using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene, Agilent, UK) in accordance with the manufacturer's protocol (primer sequences available on request). The variant c.532A>G (p.Ile178Val) was investigated in isolation and introduced within the color-normal haplotype minigenes LVAIS or MVAIA using the same protocol. XL10-Gold ultracompetent cells (Agilent Technologies, Waldbronn, Germany) were transformed with 20 ng of minigene DNA and plasmid DNA was isolated using a QuickLyse Miniprep kit (Qiagen, Crawley, UK). All constructs were sequenced to ensure fidelity and orientation.
HEK293 cells were transfected with 500 ng of wild-type or mutant minigenes using Lipofectamine (Life Technologies Ltd, Paisley, UK). After 24 hr incubation, cells were harvested and lysed using 600 μl lysis buffer. Total RNA was extracted using an RNeasy mini kit (Qiagen, Crawley, UK) with on-column DNAse treatment (Promega, Hampshire, UK), according to the manufacturer's instructions. cDNA was synthesized from approximately 1 μg total RNA using a Tetro cDNA Synthesis Kit (Bioline Reagents Ltd, London, UK) with TetroRTase enzyme and Anchored Oligo (dT) primers in accordance with the manufacture's guidelines. Vector-specific primers were used to establish cDNA linearity loading controls for the experimental PCR assays [Ueyama et al., 2012]. The products were analyzed on a 2% agarose gel and either sequenced directly, or gel extracted using a QIAquick® gel extraction kit (Qiagen) and cloned into pGEM®-T Easy Vector System (Promega) prior to sequencing. Plasmid DNA was extracted using the GenElute Plasmid Miniprep Kit (Sigma–Aldrich Company Ltd., Dorset, UK), followed by sequencing using the Big Dye Terminator v3.1 Cycle Sequencing reagents as recommended by the manufacturer (Applied Biosystems).
Databases and Bioinformatics
Ensemble and UCSC Genome Browsers were used for analyses of the Human Feb 2009 (GRCh37/hg19) Assembly. In silico splicing analyses were performed using pre-mRNA splicing prediction programs RESCUE-ESE [Fairbrother et al., 2002; Fairbrother et al., 2004] and FAS-ESS [Wang et al., 2004]. Candidate exonic splicing enhancer (ESE) sequences in exon 3 of the opsin gene haplotypes were identified using the human ESE parameter of RESCUE-ESE (http://genes.mit.edu/burgelab/rescue-ese/). The FAS-hex2 set of FAS-ESS (http://genes.mit.edu/fas-ess/) enabled detection of candidate exon splice silencer motifs in each exon 3 haplotype.
Statistical Analysis
The relationship between flicker ERG amplitudes (corneal recordings) and patient age was examined in the interchange haplotype group by linear regression and a correlation coefficient was determined using Microsoft Excel. The coefficient of variation (r2) was derived from the linear regression through six data points. A P value was obtained using a table of critical values of the t-distribution for n-2 degrees of freedom.
Results
Clinical data for 26 affected males from 22 different families, with an age range of 4–80 years, are summarized in Supp. Table S1. All pedigrees are illustrated in Fig. 1, with the exception of Family 22, which is shown in Supp. Figure S1. For ease of interpretation, clinical and genetic detail of individuals within these pedigrees have been assigned subject numbers (S1–S26) and are annotated on Figs. 1 and 2, Supp. Figure S1, Supp. Table S1, and all clinical figures.
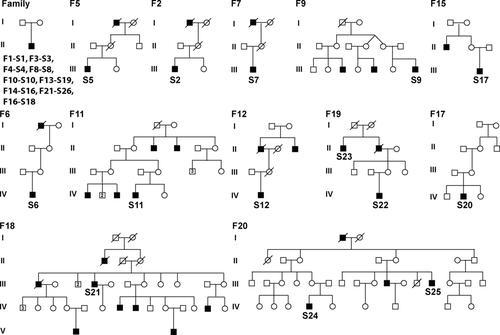

A diagrammatical representation of the cone opsin gene array structure in each family is shown in Fig. 2, classified into three distinct mutation categories: (1) deletion of the LCR (five families), (2) single hybrid gene with inactivating missense mutation c.607T>C (p.Cys203Arg) (eight families), and (3) SNP interchange haplotypes (nine families).
All subjects, with the exception of subject S26, reported onset of symptoms in their first decade of life with impaired color and central vision being the main symptoms. S26 reported normal vision until his late 20s, when he noticed reduced central vision. All patients were ametropic, most commonly myopic.
Deletion of the LCR and Associated Phenotypes
Deletions of the LCR were identified in five unrelated male probands (S1–S5) (Figs. 1 and 2; Supp. Table S1). The deletion in S4 involved the LCR-core sequence and the first upstream STS (7.5 kb) but not the upstream promoter or coding regions of the array. Three probands (S1, S2, and S5) had deletions of the LCR-core region that extended upstream to include the first and second STSs (7.5 and 8.4 kb) and downstream into the coding sequence of the proximal L-gene (Fig. 2). Genomic DNA of S3 did not amplify for regulatory regions, STSs, or cone opsin exons, which indicated the presence of an extensive deletion involving the entire opsin array. The first exons of the two genes flanking the opsin array, MECP and TKTL, were successfully amplified in this individual, indicating these genes were not deleted (Fig. 2).
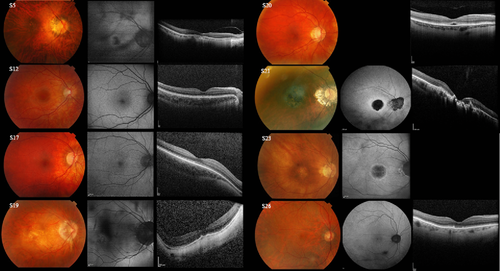
The five probands with LCR deletions had ages ranging from 10–58 years and had onset in early infancy. All five had low to high myopia with reduced central visual acuity ranging from 6/30 to 6/60 (Supp. Table S1). Four out of five subjects (S1–S4, aged 10–17 years) had nystagmus, whereas S5 (age 58 years), who was first seen in adulthood in his late 40s, did not have documented nystagmus. Fundus examination revealed myopic fundi. Macular SD-OCT showed disruption of the inner segment ellipsoid (ISe) layer to a greater extent in S5 (age 58 years) compared with S2 (age 14). Follow-up of these subjects from 2–8 years showed no evidence of clinical progression. Figure 3 shows the myopic fundus photograph, abnormal central FAF, and disrupted ISe SD-OCT image of S5. Two children underwent ERG testing with surface electrodes (S1 and S3) and two subjects (S2 and S5) underwent ERGs at the ages of 10 and 51 years (Supp. Tables S2 and S3 and Supp. Figs. S2–S4). All had undetectable or severely abnormal flicker and/or photopic ERGs consistent with severe generalized cone system dysfunction. All four also had undetectable PERGs, consistent with severe macular dysfunction. Scotopic ERG showed mild rod system involvement in S2 only, with a preserved S-cone ERG.
Hybrid Genes with an Inactivating p.Cys203Arg Missense Mutation and Associated Phenotypes
A hemizygous c.607T>C, p.Cys203Arg missense mutation (Fig. 2) was identified in eight probands (S6–S12 and S13), and two affected relatives of S13 in Family 22 (S14 and S15) (Fig. 1; Supp. Fig. S1). Each subject had a single L-/M-hybrid gene sequence with the inactivating p.Cys203Arg missense mutation in exon 4. No wild-type cone opsin exon 4 sequence was identified.
In Family 22, the proband S13 (IV-1) aged 27 years, his affected half-brother S14 (IV-3), and nephew S15 (V-2) were diagnosed with “X-linked cone dystrophy” (Supp. Fig. S1). S13 had a bull's-eye maculopathy that had been noted from the age of 10 years. All three affected subjects had macular pigmentation that had been visible from early childhood. Interestingly, the maternal grandfather (III-4) and two half-brothers (V-1 and V-3) of the youngest affected individual S15 (V-2) had red/green color blindness but had no significant loss of visual acuity, myopia, or nystagmus (Supp. Fig. S1). Unusually, the mother of S15 (IV-7) also had red/green color blindness. Analysis of the opsin gene array in this family showed segregation of two different opsin genotypes with the two different phenotypes (Supp. Fig. S1). The three affected visually impaired males (Supp. Table S1; patients S13–S15) had a single L-/M-hybrid gene with the inactivating p.Cys203Arg mutation, inherited from their respective mothers (Supp. Fig. S1). The half-brothers (V-1 and V-3) with red/green color blindness had a cone opsin array comprising only a single functional M-gene accounting for the color vision deficiency (protanopia). Their color blind mother (IV-7) was a carrier of the single hybrid mutant p.Cys203Arg on one X-chromosome, and a single M-gene on the other (inherited from her father; Supp. Fig. S1).
Individuals in the study with this mutation class had onset in early infancy. The age range of affected individuals was 4–27 years with visual acuity ranging from 6/19 to 3/60. Nine of 10 subjects had documented nystagmus and all had low-to-moderate myopia or myopic astigmatism. S11 (age 16 years) had some peripheral hyperpigmentation, and S13 and S15 from Family 22 (age 27 and 19 years) had macular atrophy, but fundus examination was unremarkable in the remaining patients. SD-OCT for S10 (age 14 years) revealed mild granular disruption of the ISe layer at the macula, but was unremarkable in the remaining three individuals that were imaged. Fundus photographs, FAF, and SD-OCT images of S12 (normal imaging) are shown in Fig. 3. In two young patients with high myopia (S10 and S12), photopic ERGs were consistent with severe generalized cone system dysfunction (Supp. Tables S2 and S3; Supp. Figs. S2–S4). The S-cone ERGs were preserved in S10 (excluded in S12 due to eye movement artifact). Scotopic ERGs were normal (S10) or mildly abnormal (S12). There was PERG evidence of severe macular dysfunction in S10 and S8, and moderate macular dysfunction in S12 (the only patient in the entire cohort to have a detectable PERG P50 component). Two subjects (S13 and S10) were followed up for 7 and 10 years, respectively, with no definite clinical evidence of progression.
Disease-Causing SNP Interchange Haplotypes and Associated Phenotypes
Nine probands (S16–21, S23, S24, and S26) had L-/M-interchange genotypes (Supp. Table S1; Figs. 1 and 2). For ease of reference and consistency with the literature, genotypes are referred to by their exon 3 protein coding haplotype (Table 1). Analysis of these gene arrays revealed the presence of two or more genes with a deleterious haplotype associated with an upstream or downstream promoter (Fig. 2).
| Exon 3 haplotype (translated) | Expected opsin function | DNA variant position and corresponding amino acid | Opsin transcript(s) | Predicted effect on protein | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
||||
| LVAIS | Normal | G |
|
G |
|
G |
|
|
|
r.[ = ] | p.[ = ] |
| MVAIA | Normal | A |
|
C |
|
G |
|
|
|
r.[ = ] | p.[ = ] |
| LIAVA | Affected | G |
|
G |
|
T |
|
|
|
r.[410_578del] | p.{Ile138Thrfs*6} |
| LVAVA | Affected | G |
|
G |
|
G |
|
|
|
r.[ = , 410_578del, 350_578delins579–127_579–1] | p.{ = , Ile138Thrfs*6, Tyr118Serfs*5} |
| MIAVA | Affected | A |
|
C/G |
|
T |
|
|
|
r.[ = , 410_578del, 350_578delins579–127_579–1] | p.{ = , Ile138Thrfs*6, Tyr118Serfs*5} |
Four individuals (S18, S19, S20, and S21) had arrays comprising only an L- or L-/M-hybrid gene with an interchange haplotype in exon 3 of either LIAVA (S21) or LVAVA (S18, S19, and S20) (Fig. 2). Five subjects (S16, S17, and S24–S26) had multigene arrays that included at least one gene with a rare interchange haplotype and at least one (M-)gene with an MVVVA haplotype. Two of these subjects (S16 and S17) had an LIAVA haplotype and one (S26) had an LVAVA haplotype. The two remaining related subjects (S22 and S23) with multigene arrays had genes encoding LIAVA and MIAVA. The interchange haplotypes in these families segregated with the condition (Supp. Table S1; Figs. 1 and 2). These data identify interchange haplotypes as a relatively common cause of disease in our cohort.
Subjects S16–26 harbored exon 3 SNP interchange haplotypes (Fig. 2) and had moderate-to-high myopia (>6 diopters), except S23 and S26 who had low myopia (<3 diopters). Onset of disease was in infancy, apart from S26 who presented in his early 20s. A greater degree of phenotypic heterogeneity was observed in this mutation class compared with the LCR deletion and inactivated single gene array classes. Visual acuity ranged from 6/12 in the younger subjects, to 6/60 in S21 and S23, who were aged 73 and 68 years, respectively. Three subjects out of eleven (S22–S24) had documented nystagmus. Fundus findings appeared to be related to patient age, with younger patients (<40 years, S16–S18, S22, and S24) showing only myopic changes or mild pigmentary abnormalities (Fig. 3), whereas all older patients (≥40 years), except S25, had evidence of macular abnormalities on examination, photography, FAF, or SD-OCT. Subject S19 (age 40 years; Fig. 3) had central outer retinal atrophy on photography and SD-OCT; subject S20 (age 49 years; Fig. 3) had subfoveal lucencies on SD-OCT; S21 (age 73 years; Fig. 3) had bilateral bull's-eye macular lesions and atrophy on FAF and SD-OCT; S23 (age 68 years; Fig. 3) also had bilateral bull's-eye lesions on FAF; and S26 (age 80 years; Fig. 3) had disruption of the outer retinal layers on SD-OCT. Subject S25 (age 51 years) was the only patient over the age of 40 years in this group with exon 3 SNP interchange haplotypes who did not have documented macular abnormalities, but he did not have FAF or SD-OCT imaging performed. ERGs performed >20 years ago in this patient showed severe cone and rod system dysfunction. In the six patients with L-/M-interchange genotypes for whom electrophysiology was performed, there was moderate-to-severe photopic flicker ERG reduction (Supp. Figs. S2–S5). There was a trend for ERG amplitudes (corrected for age) to decrease with increasing age (Supp. Fig. S4). S-cone ERG amplitudes were normal in all but one subject (S23; age 63 years). Scotopic red flash ERGs revealed no detectable dark-adapted cone system function in any patient (Supp. Fig. S2) and PERGs were undetectable in six of six cases. Scotopic ERGs were normal (S16–S18, S21, and S23) or had some mildly (S20 and S22) or moderately (S19 and S25) abnormal parameters (Supp. Figs. S2 and S3). The oldest patient (S26; 69 years at the time of first ERG) showed evidence of marked reduction in scotopic ERGs when tested 5 years later (Supp. Fig. S5); patient 21 (59 years at time of ERG) showed an increase in rod (DA 0.01) ERG timing by 15 msec after 12 years (Supp. Fig. S5).
In Vitro Splicing Assay and In Silico Analysis
An in vitro splice assay was performed to test the effects of the opsin interchange haplotypes as a result of combinations of SNP variants associated with disease. The splicing patterns of two color-normal wild-type opsin haplotypes LVAIS (L-) and MVAIA (M-) were compared with three haplotypes encoding LIAVA, LVAVA, and MIAVA that were associated with disease, and therefore presumed loss of functional opsin (Supp. Table S1; Table 1). An analysis of the resulting spliced cDNA products demonstrated that the wild-type and mutant constructs produced differently spliced transcripts (Table 1; Fig. 4).
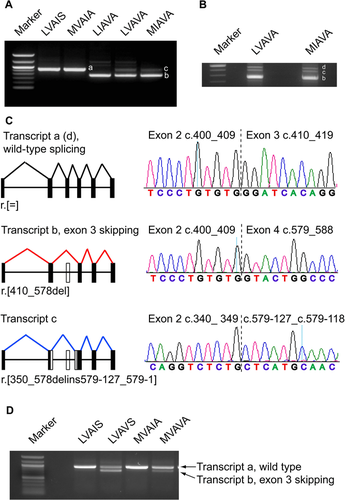
The wild-type constructs produced a single product corresponding to the correctly spliced cone opsin transcript. In contrast, LIAVA produced a single smaller transcript due to exon 3 skipping (exon 2 spliced onto exon 4), as previously described [Ueyama et al., 2012]. Interestingly, LVAVA and MIAVA resulted in a different pattern of aberrant splicing, with several transcripts being produced (Fig. 4). Cloning and sequencing revealed a transcript with exon 3 skipping, as established for the LIAVA haplotype, an additional aberrant transcript with partial exon 2 spliced on to intron 3, and a low level of the correctly spliced cone opsin transcript (Fig. 4).
Given that all three disease-associated interchange haplotypes tested resulted in aberrant splicing, pre-mRNA splicing prediction programs were used to assess whether the SNPs could abolish or generate splice donor, splice acceptor, exonic splice enhancer (ESE) or exonic splice silencer (ESS) sites. The three interchange haplotypes were not predicted to introduce any cryptic splice donor or acceptor sites, or to abolish the normal exon 3 donor and acceptor sites. RESCUE-ESE predicted that four ESEs are present in wild-type OPN1LW sequence and nine in OPN1MW [Fairbrother et al., 2002] (Supp. Fig. S6). Interestingly, the variant c.532A>G present in all three interchange haplotypes resulted in loss of an ESE, whereas the other variants in the haplotypes had no predicted effect (Supp. Fig. S6). Similarly, FAS–ESS predicted that 10 ESSs are present in OPN1LW and OPN1MW wild-type sequence [Wang et al., 2004]. The c.532A>G variant resulted in the generation of an additional splice silencer at this site, whereas the other variants had no effect (Supp. Fig. S6). This analysis predicted that the c.532A>G variant present in all disease-associated haplotypes could have a major role in the aberrant splicing observed in the in vitro splicing assay. The effect of altering this single nucleotide was therefore investigated in isolation in the color-normal wild-type L- and M-genes. The data show that the c.532A>G variant on a wild-type haplotype is sufficient, in isolation, to cause aberrant splicing (exon 3 skipping) in both the L-and M-opsin genes (Fig. 4).
Discussion
This study describes detailed phenotypic and genetic findings of one of the largest cohorts to date of patients harboring mutations in the cone opsin gene array and expands our knowledge of the genetic mechanisms and phenotypic variability.
There was considerable variation in the severity and progression of disease in the cohort, with some evidence of genotype–phenotype correlation. Subjects with LCR deletions and those harboring the p.Cys203Arg variant were more likely to have nystagmus, poor vision, and severe cone system dysfunction. These patients generally had normal fundus imaging and no evidence of progression with age, the exception being some individuals with p.Cys203Arg mutations who had evidence of retinal degeneration. Rod system involvement varied with a young patient from each of these two patient groups showing mild abnormalities of the scotopic dim flash (DA 0.01) and bright flash (DA 11.0) ERGs. It is noted that reduction in bright flash ERG (Supp. Tables S2 and S3) could partly relate to a loss of the dark-adapted cone system contribution, also absent in the scotopic red flash ERGs (Supp. Fig. S2). However, the DA0.01 ERGs are recorded to a dim flash, which is below cone system threshold and is selective for the rod system.
In contrast, patients with exon 3 interchange haplotypes were less likely to have nystagmus and had better VA, particularly below the age of 40. The phenotype appeared age dependent, with patients below the age of 40 having normal fundus appearance and imaging, whereas all patients over the age of 40 years showed marked central macular abnormalities on fundus examination, FAF, and SD-OCT, with the exception of S25, who had incomplete imaging. All interchange haplotype patients had evidence of cone system dysfunction with a tendency for flicker ERG amplitudes to be smaller with increasing age (Supp. Fig. S4). Additionally, serial ERGs in two of the oldest patients suggested progressive rod system dysfunction in the advanced stages of disease. S-cone ERG amplitudes were abnormal in only one of the oldest patients. These findings suggest that patients with LCR deletions are more likely to have a typical BCM phenotype that is predominantly nonprogressive, although progression has previously been reported in some cases with LCR deletions [Fleischman and O'Donnell, 1981; Nathans et al., 1989; Ayyagari et al., 1999]. Patients with p.Cys203Arg missense variants can also have a typical BCM phenotype or experience slow progression over time.
In our cohort, patients with exon 3 interchange haplotypes commonly had a progressively worsening phenotype, including visual acuity reduction, with onset of macular atrophy after the age of 40 years, and electrophysiological evidence of progressive loss of cone function with increasing rod involvement over time. These results are in keeping with other reports showing that some patients with exon 3 interchange haplotypes have more severe outer retinal atrophy than patients with p.Cys203Arg mutations, and that these patients also report a progressive loss of visual function [Carroll et al., 2012]. The interchange haplotype patients in the cohort, except S26 (who harbors the LVAVA haplotype), all developed symptoms in their first decade. Importantly, high-resolution adaptive optics imaging has shown differences in the cone mosaic in patients with p.Cys203Arg and exon 3 interchange haplotypes [Carroll et al., 2009, 2012] as well as in carrier females with LCR deletions, further highlighting the phenotypic differences of each mutation category.
We observed three different exon 3 SNP interchange haplotypes encoding LIAVA, LVAVA, and MIAVA that were associated with disease in our cohort. Four families had multigene arrays that comprised a third, common M-gene haplotype, MVVVA [Neitz et al., 2004]. It is evident that both MIAVA and LVAVA result in aberrant pre-mRNA splicing in vitro that is different to that caused by LIAVA. The LIAVA haplotype transcript resulting from exon 3 skipping is predicted to be degraded by nonsense-mediated decay (NMD) or to result in a frameshift, producing a truncated protein with a premature stop codon (p.Ile138Thrfs*6). In comparison, the MIAVA and LVAVA haplotypes both resulted in an additional aberrant transcript that is also predicted to be degraded by NMD or to produce an alternative frame-shifted truncated protein product (p.Tyr118Serfs*5). However, a residual level of normally spliced opsin was also detected, at much lower levels than the aberrant transcripts. It is plausible that these residual levels of correctly spliced cone opsin in individuals with an LVAVA or MIAVA haplotype could result in a population of cones that express some functional opsin. Over time, the survival of these photoreceptor cells may be impaired by the simultaneous expression of the aberrant transcripts or mutant proteins, if expressed. Individuals with an LIAVA haplotype may be predicted to have congenital disease and a more stable disease course. However, this study is limited by the relatively small cohort size in each category and there is inherent difficulty in extrapolating these data to draw conclusions on genotype–phenotype correlation. Further detailed serial quantitative high-resolution retinal imaging in larger cohorts of molecularly confirmed patients will be necessary to establish the cellular phenotype associated with the different classes of cone opsin mutations. Robust data on their natural history will help to identify which patients/genotype classes may be potentially most amenable to novel interventions in the future.
The c.532A>G variant was common to all three disease-associated interchange haplotypes. We have shown that c.532A>G alone can cause aberrant pre-mRNA splicing in vitro, suggesting it contributes to mis-splicing in all three haplotypes and that the other seven SNPs in the haplotypes further modulate its functional outcome. The minor allele “G” of c.532A>G (ref dbSNP rs145009674) has a reported minor allele frequency of 2.2% in both the 1000 Genomes Dataset and the NHLBI Exome Sequencing Project (ESP). However, these data do not take into account the position or number of L- or M-genes in the array. It would be interesting to investigate the frequency, context, and position of this SNP in red/green color blind subjects, who account for up to 8% of males and 0.5% of females among populations of Northern European origin [Deeb and Motulsky, 2011], and in color vision normal subjects.
Acknowledgments
We thank all the families for participating in this study, and we are grateful to Joseph Carroll for critical evaluation of this manuscript.
Disclosure statement: The authors declare no conflict of interest.




