Mutation in The Nuclear-Encoded Mitochondrial Isoleucyl–tRNA Synthetase IARS2 in Patients with Cataracts, Growth Hormone Deficiency with Short Stature, Partial Sensorineural Deafness, and Peripheral Neuropathy or with Leigh Syndrome
Communicated by Ming Qi
Contract grant sponsors: Government of Canada through the Canadian Institutes of Health Research (EAS); Genome Canada; Genome Quebec; Genome British Columbia; the Ontario Genomics Institute (OGI-049); Centre de Recherche du CHU Ste-Justine.
ABSTRACT
Mutations in the nuclear-encoded mitochondrial aminoacyl–tRNA synthetases are associated with a range of clinical phenotypes. Here, we report a novel disorder in three adult patients with a phenotype including cataracts, short-stature secondary to growth hormone deficiency, sensorineural hearing deficit, peripheral sensory neuropathy, and skeletal dysplasia. Using SNP genotyping and whole-exome sequencing, we identified a single likely causal variant, a missense mutation in a conserved residue of the nuclear gene IARS2, encoding mitochondrial isoleucyl–tRNA synthetase. The mutation is homozygous in the affected patients, heterozygous in carriers, and absent in control chromosomes. IARS2 protein level was reduced in skin cells cultured from one of the patients, consistent with a pathogenic effect of the mutation. Compound heterozygous mutations in IARS2 were independently identified in a previously unreported patient with a more severe mitochondrial phenotype diagnosed as Leigh syndrome. This is the first report of clinical findings associated with IARS2 mutations.
Mitochondrial function is dependent on both the mitochondrial genome and the expression of over 1,000 proteins encoded by the nuclear genome, which are synthesized in the cytoplasm and imported into mitochondria [Florentz et al., 2003]. The mitochondrial respiratory chain is composed of 13 mitochondrial translation products (subunits) that are encoded in the mitochondrial genome, complemented by 72 additional subunits of nuclear origin [Edvardson et al., 2007]. Translation of mitochondrial-encoded proteins is dependent on aminoacyl–tRNA synthetases that are themselves all encoded by the nuclear genome, translated in the cytoplasm, and transported into the mitochondria. Defects in the nuclear genes encoding these tRNA synthetases have emerged as a new group of human mitochondrial diseases, presenting striking tissue-specific phenotypes and surprisingly not always leading to obviously impaired mitochondrial protein synthesis or oxidative phosphorylation system (OXPHOS) [Konovalova and Tyynismaa, 2013].
Here, we define a novel recessive disorder CAGSSS, seen in one new index case and two previously reported cases [Liberfarb et al., 1993]. Clinical follow-up of the earlier cases, and ascertainment and phenotyping of the third patient, led us to propose this novel disorder nomenclature based on the presence of cataracts (CA), growth hormone deficiency (G), sensory neuropathy (S), sensorineural hearing loss (S), and skeletal dysplasia (S).
The novel index case (case 1), a female now 32 years old, was born to healthy French-Canadian parents after an uncomplicated pregnancy; the parents are first cousins (Fig. 1A). Her developmental motor milestones were mildly delayed; she sat unaided at age 7 months and walked at age 19 months. Mildly dysmorphic facial features include thick eyebrows, a prominent forehead, and a small mouth, nodules of the proximal interphalangeal joints of the hands, mild thoracal scoliosis, and genu valgum (Fig. 1B–D). She had overall significantly reduced growth during infancy and childhood (Supp. Fig. S1). At 15 years, she was seen in Endocrinology for severe short stature and hypoglycaemia, found to be due to growth hormone deficiency together with a presumed cortisol deficiency of central origin. A short course of GH therapy was given and she showed a clear growth response (Supp. Fig. S1). MRI showed an atrophied pituitary adenohypophysis and a small neurohypophysis, consistent with growth hormone deficiency. She had her menarche at age 14 with subsequently normal cycles. Apart from occasional fasting hypoglycemia, which has improved with growth hormone treatment, her metabolic status remains normal. Cataracts were observed beginning around the age of 17 months (not surgically corrected until age 22 years); bilateral hearing loss was observed beginning around the age of 2 years (corrected with hearing aids); distal sensory neuropathy involving loss of sensitivity to pain, temperature, and touch was observed from the age of 9.5 years; and multiple skeletal anomalies beginning with bilateral hip dislocation were observed first at the age of 2 years (Supp. Fig. S2) with a diagnosis of spondylo-epiphyseal dysplasia. A recent spine survey identified diffuse cervical spinal stenosis and confirmed the hypoplasia of C2-odontoid, with suspicion of cervical spine instability (Fig. 1E). Recently, she has also developed severe difficulties swallowing solids, and a type II achalasia of her oesophagus was diagnosed. No evidence of obstruction was seen on an upper GI series, and the patient is awaiting surgery on the gastrooesophageal sphincter. She continues to report an overall improvement including increased sense of well-being and exercise tolerance. She has no cognitive deficits, is alert and engaged, and she currently pursues university studies and is living independently in a group home for physically handicapped individuals. See Supp. Methods for additional clinical details.
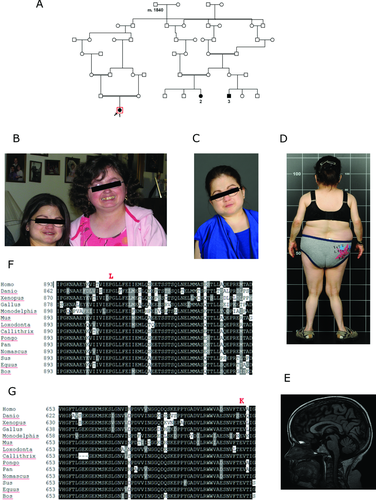
The patient is phenotypically very similar to two individuals (cases 2, 3; Fig. 1A) previously reported in the literature with an overlapping suite of clinical features [Liberfarb et al., 1993]. The other two patients share the same eye, cochlear, neurologic, and endocrine findings, with short-stature secondary to growth hormone deficiency, and hip dysplasia with scoliosis (Supp. Table S1). These two patients are first cousins to each other, with documented parental consanguinity in one case. They are also of French-Canadian descent, and are genealogically related to the new index case (Fig. 1A). The female cousin, though older, noticeably resembles the new proband (Fig. 1B).
Hypothesizing that our cases might represent a novel recessive disorder with a possible founder effect of French-Canadian origin, we performed homozygosity mapping using high-density SNP genotyping and whole-exome sequencing in parallel (see Supp. Methods for details). The molecular analysis was carried out in the context of the Canadian national FORGE project on rare pediatric disorders [Beaulieu et al., 2014]. Based on SNP genotyping, the three patients shared one extended region of homozygosity; a 2.35 Mbp region from SNP rs12728902 to rs10489954 on chromosome 1 (Supp. Fig. S3A and B). Homozygosity Mapper does not require identity-by-state among multiple samples, only overlapping homozygosity, however, haplotype analysis confirmed that all samples were homozygous for the same allele at each SNP in the candidate region. The minimum interval of shared homozygosity contains 15 annotated protein-coding genes as well as several microRNAs (Supp. Table S1).
In parallel, we carried out whole-exome sequencing on all three patients. After filtering to exclude common variants (those with a minor allele frequency >1% in 1000 Genomes) and potential technical artifacts of sequencing chemistry or short-read alignment and annotation (variants present in >10 control exomes analyzed locally from unrelated projects), and after removing known artifact-prone gene families (HLAs, MUCs, MAGEs, NBPFs, and PRAMEFs), there were 2,760 rare variants in protein coding or adjacent intronic regions among all three exomes, 1,235 excluding synonymous (i.e., silent) variants. Of these, only nine variants were observed simultaneously in all three exomes (Supp. Table S2). Only one variant among these was homozygous in all three exomes, and in the shared homozygous region defined by SNP genotyping. This was also the only rare nonsynonymous variant homozygous in all three exomes using the less stringent filters of <3% in 1000 Genomes and <30 control exomes. This was a proline to leucine change at position 909 (c.2726C>T, p.P909L) in the gene IARS2, a nuclear gene encoding mitochondrial isoleucyl–tRNA synthetase (MIM #612801) (Supp. Fig. S4A and B). We confirmed segregation of the shared variant by Sanger sequencing on both parents of one of the first cousins, and one parent of the other; these parents were heterozygous for this variant, as was one unaffected sibling (Supp. Fig. S4C, data not shown). The variant was not seen in 540 control exomes (1,080 chromosomes) that include French-Canadian individuals with unrelated clinical conditions. No other gene was potentially compound heterozygous carrying two different rare variants in all three exomes, thus IARS2 was the only gene consistent with potentially pathogenic recessive variation shared among the three patients.
The IARS2 gene product is annotated as a mitochondrial protein by the human mitoproteome project [Cotter et al., 2004]. Position 909 lies in the predicted anticodon RNA-binding domain of class I aminoacyl–tRNA synthetases (InterPro domain IPR009080, SCOP superfamily SSF47323). The gene IARS2 is highly conserved evolutionarily, and we were able to identify and compare orthologs from multiple species showing that proline 909 is completely conserved in aligned vertebrates (Fig. 1F). The variant p.P909L was predicted to affect protein function by SIFT (Supp. Fig. S5A), to be probably damaging by PolyPhen2 HumDiv and possibly damaging by PolyPhen2 HumVar (Supp. Fig. S5B), to be deleterious by Provean with a score of −4.9 (−2.5 or below is deleterious threshold) (Supp. Fig. S5C), to be disease causing by MutationTaster (score greater than 0.999 hence high-security prediction, with simple_aee model), to be likely to interfere with function by Align-GVGD (GD score 97.78, class C65), but to be neutral by SNPs&GO (reliability index 6 on a scale of 0–10, 10 is most reliable) and tolerated by FATHMM.
Further evidence that the p.P909L mutation in IARS2 affects gene activity came from direct analysis of IARS2 protein in patient skin cells. We obtained primary skin fibroblasts from one affected patient carrying this mutation, which were subsequently immortalized [Yao and Shoubridge, 1999]. Protein extracts of these cells were analyzed by immunoblotting with a polyclonal antibody against human IARS2. There was a significant reduction in IARS2 protein in these cells compared with wild-type control fibroblasts (Fig. 2A, black arrow). To test whether the p.P909L mutation in IARS2 affected translation of mitochondrial-encoded proteins, we pulse-labeled mtDNA-encoded polypeptides in patient and control fibroblasts [Sasarman and Shoubridge, 2012], but did not detect any difference (Fig. 2B). Similarly, Blue-Native polyacrylamide gel electrophoretic analysis of oxidative phosphorylation complexes [Leary, 2012] did not show any obvious difference between patient and control fibroblasts (Fig. 2C).

The genetic findings, combined with the reduced IARS2 protein levels in cells derived from one patient, strongly argue that the variant in IARS2 is a pathogenic mutation and the primary cause of the phenotype in these three related patients. Additional functional experiments to investigate the impact of the reduced levels of IARS2 protein in humans, such as aberrant translation of the OXPHOS respiratory chain and concomitant production of ROS, are ideally addressed in tissue biopsies that are specific to the phenotype, which are in this case unavailable. As reported in previous publications on mtARS diseases, immortalized patient fibroblast cell lines typically do not show a measurable OXPHOS complex deficiency on protein gels, even in severely affected cases [Konovalova and Tyynismaa, 2013].
In the course of our study, we became aware of an additional, unrelated case carrying mutations in the IARS2 gene. This patient (case 4), not previously reported, is from the Western United States, is unrelated to the three CAGSSS patients and is reportedly of Scandinavian Caucasian ethnicity. He was initially ascertained at 4 weeks of age due to an episode of severe cerebral degeneration. He was eventually diagnosed with Leigh syndrome (MIM #256000), a genetically heterogeneous subset of mitochondrial disorders, based on the location of stroke in the thalami and basal ganglia, severe skeletal myopathy, in particular ocular myopathy, and laboratory testing (see Supp. Methods for details). The patient was managed with cofactor therapy (combined vitamins) and physical therapy, but died at 18 months of age due to complications. The patient's parents are healthy. The patient was referred to Courtagen Life Sciences, Inc. for next-generation sequencing of a panel of 1,092 genes with potential relevance to mitochondrial disorders. The only pathogenic variants were two rare variants in IARS2: c.1821G>A, p.W607X, and c.2122G>A, p.E708K (Supp. Fig. S6A and B). The two variants were transmitted from different parents (stop codon paternal, missense maternal, data not shown), indicating that the patient is a compound heterozygote. The first mutation, c.1821G>A, p.W607X, truncates the protein removing 405 amino acids and is expected to be severely pathogenic. The second mutation, c.2122G>A, p.E708K, is at the junction of the catalytic core domain and the anticodon-binding domain; the residue is highly conserved among vertebrates (see Fig. 1G) but less well in lower organisms for which the alignments are also less certain. The p.E708K variant is predicted to be damaging by SIFT (Supp. Fig. S7A), to be possibly damaging by PolyPhen2 HumDiv and benign by PolyPhen2 HumVar (Supp. Fig. S7B), to be disease causing by MutationTaster (score greater than 0.999 hence high-security prediction, with simple_aee model), to be disease causing by SNPs&GO (reliability index 4), to be somewhat likely to interfere with function by Align-GVGD (GD score 56.87, class C55), but to be neutral by Provean (score −2.39, slightly above the cutoff of −2.5) (Supp. Fig. S7C), and tolerated by FATHMM. The p.E708K variant is present in dbSNP (rs143722284), which reports it as seen twice by 1000 Genomes (one genome, one exome), once in the NHLBI exome sequencing project, and once (not validated) on an exome chip; presumably, these are all heterozygous alleles. The more severe phenotype observed in this patient compared with the three previous patients is likely due to one of the two IARS2 mutant alleles being a stop codon, presumably generating complete loss of function of that allele.
All three variants in IARS2 have been submitted to ClinVar and assigned preliminary accession numbers pending database curation: c.1821G>A SCV000148127; c.2122G>A SCV000148128; c.2726C>T SCV000148129.
Based on combined biochemical and bioinformatic analyses, the human nuclear genome encodes 19 mitochondrial–tRNA synthetases [Bonnefond et al., 2005]. These genes are different than their counterparts encoding cytoplasmic enzymes, with the exception of two enzymes (glycyl– and lysyl–tRNA synthetases) for which the mitochondrial and cytoplasmic versions are encoded by alternative isoforms of two respective genes. In addition, the nuclear genome does not appear to encode a mitochondrial-localized glutaminyl–tRNA synthetase; as in these bacteria the appropriate mitochondrial tRNAgln is first charged with glutamate via a glutamyl–tRNA synthetase followed by amidation of the aminoacyl group to generate the correctly charged gln-tRNAgln [Nagao et al., 2009]. Mutations in other of these genes yield a range of clinical phenotypes, only partly overlapping with our cases and without universal features (Supp. Table S3 and recent review) [Konovalova and Tyynismaa, 2013]. In all cases, the conditions are recessive with homozygosity or compound heterozygosity in affected family members; further, most reported mutations are missense or intronic, suggesting that complete loss of any of these tRNA synthetases might be lethal. In one recent study, patients with explicit OXPHOS disease were sequenced for the mitochondrial genome plus approximately 1,000 candidate nuclear genes known or predicted to play a role in mitochondrial function. Among others, candidate pathogenic mutations were found in two of the mitochondrial–tRNA synthetases (EARS2 and AARS2) [Calvo et al., 2012]. Surprisingly, mutation of the mitochondrial genome-encoded gene for the isoleucyl tRNA itself, MTTI (MIM #590045), causes a different phenotype than the CAGSSS or Leigh syndrome phenotypes reported here for the isoleucyl tRNA charging enzyme, specifically hypertrophic cardiomyopathy [Perli et al., 2012]. Variable penetrance among family members sharing the causal MTTI mutation was associated with differential expression levels of the charging enzyme IARS2. The phenotypes in our three related French-Canadian patients have multiple features not yet reported for mutations in other mitochondrial tRNA synthetases, particularly cataracts, growth hormone deficiency, skeletal changes, and achalasia (Supp. Table S3). However, these have been noted as features of mitochondrial disorders either in general or specifically for other genes [Chelimsky et al., 2005; Garcia-Cazorla et al., 2005; Wolny et al., 2009; Gronlund et al., 2010; Schaefer et al., 2013; Mehawej et al., 2014] Thus, we have significantly expanded the phenotypic range for this class of important mitochondrial genes.
Acknowledgments
Foremost, we thank the families who generously contributed their time and materials to this research study and Dr. Ruth Liberfarb for her valuable help in contacting some of the families. Exome sequencing and SNP genotyping was performed at the Genome Quebec and McGill University Centre for Innovation, or at Courtagen Life Sciences, Inc. The FORGE Canada Consortium: Finding of Rare Disease Genes in Canada; Steering Committee consists of Kym Boycott (leader; Ottawa, Ontario), Jan Friedman (coleader; Vancouver, British Columbia), Jacques Michaud (coleader; Montreal, Quebec), Francois Bernier (Calgary, Alberta), Michael Brudno (Toronto, Ontario), Bridget Fernandez (St. Johns, Newfoundland), Bartha Knoppers (Montreal, Quebec), Mark Samuels (Montreal, Quebec), and Steve Scherer (Toronto, Ontario). We also thank Janet Marcadier (Clinical Coordinator) and Chandree Beaulieu (Project Manager) for their contribution to the infrastructure of the FORGE Canada Consortium. See Supporting Information for individual author contributions to this study.
Disclosure statement: Richard Boles, Katherine Sheldon, Mariella Simon, Amir Zare, and Kevin McKernan are or were associated with Courtagen Life Sciences, Inc. Other authors state that they have no conflicts of interest.




