Online Registry for Mutations in Hereditary Amyloidosis Including Nomenclature Recommendations
Communicated by Alastair F. Brown
ABSTRACT
Hereditary systemic amyloidosis comprises a group of rare monogenic diseases inherited in an autosomal dominant fashion. It is associated with mutations in genes encoding eight different proteins, including transthyretin, apolipoprotein AI, apolipoprotein AII, lysozyme, fibrinogen A α-chain, cystatin C, gelsolin and beta-2-microglobulin. With support from the EU FP6 EURAMY project we have designed an online registry of genes and mutations in hereditary amyloidosis including their associated clinical phenotypes, with a view to having a single free online portal for the collection and distribution of this information. Users can search the registry by either mutation, phenotype or authors who have published or submitted mutations. It provides a submission form for reporting newly identified mutations. We also wanted to introduce nomenclature which complies with recommendations set out by Human Genome Variation Society and HUGO Gene Nomenclature Committee for description of new and known genetic variants. We hope this registry would be a useful and convenient tool for the medical and scientific community. ©2014 Wiley-Liss, Inc.
Introduction
Amyloidosis is a rare disorder in which normally soluble proteins undergo a conformational change to generate insoluble, toxic aggregates that are deposited in the extracellular space as abnormal β-sheet fibrils resulting in progressive disruption of the structure and function of affected tissues and organs [Pepys, 2006]. Amyloidosis is a remarkably heterogeneous disease, which can be acquired or hereditary, and systemic or localized. The commonest diagnosed form of systemic amyloidosis is AL amyloidosis, in which the fibrils are derived from monoclonal immunoglobulin light chains and consist of the whole or part of the variable (VL) domain.
Hereditary systemic amyloidosis is caused by inheritance of an abnormal gene in an autosomal dominant fashion, which leads to the life-long production of a potentially amyloid forming protein. Eight different fibril proteins have been identified with variant forms which are associated with clinically important familial amyloidosis [Sipe et al., 2012]: transthyretin (ATTR) [Saraiva et al., 1984; Benson and Uemichi, 1996], apolipoprotein AI (AApoAI) [Nichols et al., 1988; Rowczenio et al., 2011], apolipoprotein AII (AApoAII) [Benson et al., 2001], lysozyme (ALys) [Pepys et al., 1993; Gillmore et al., 1999], fibrinogen A α-chain (AFib) [Benson et al., 1993; Gillmore et al., 2009], cystatin C (ACys) [Ghiso et al., 1986], gelsolin (AGel) [Maury et al., 1992] and beta-2-microglobulin (Aβ2M) [Valleix et al., 2012] (Table 1).
| Gene Acronym | Locus | NCBI cDNA Reference Sequence | Gene/Locus MIM No | Protein | Fibril Protein Acronym | NCBI Protein Reference Sequence | Protein length | Signal Peptide/Propeptide | Mature protein | Amyloidosis Syndrome/Alternative name(s) | Phenotype MIM No |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TTR | 18q12.1 | NM_000371.3 | 176300 | Transthyretin | ATTR | NP_000362.1 | 147 AA | 20 AA | 127 AA | ATTR/Familial Amyloid Polyneuropathy | 105210 |
| APOA1 | 11q23.3 | NM_000039.1 | 107680 | Apolipoprotein AI | AApoAI | NP_000030.1 | 267 AA | 18 AA/6 AA | 243 AA | AApoAI/Ostertag Amyloidosis, Nonneuropathic Systemic Amyloidosis | 105200 |
| APOA2 | 1q23.3 | NM_001643.1 | 107670 | Apolipoprotein AII | AApoAII | NP_001634.1 | 100 AA | 18 AA/5 AA | 77 AA | AApoAII | NONE |
| FGA | 4q31.3 | NM_000508.3 | 134820 | Fibrinogen alpha chain | AFib | NP_000499.1 | 866 AA | 19 AA/16 AA | 831 AA | AFib/Hereditary Renal Amyloidosis | 105200 |
| LYZ | 12q15 | NM_000239.2 | 153450 | Lysozyme | ALys | NP_000230.1 | 148 AA | 18 AA | 130 AA | ALys/Hereditary Renal Amyloidosis | 105200 |
| GSN | 9q33.2 | NM_000177.4 | 137350 | Gelsolin | AGel | NP_000168.1 | 782 AA | 27 AA | 755 AA | AGel | 105120 |
| B2M | 15q21.1 | NM_004048.2 | 109700 | Beta-2-microglobulin | Aβ2M | NP_004039.1 | 119 AA | 20 AA | 99 AA | Aβ2M | 105200 |
| CST3 | 20p11.21 | NM_000099.3 | 604312 | CystatinC | ACys | NP_000090.1 | 146 AA | 26 AA | 120 AA | ACys/ Cerebral Amyloid Angiopathy | 105150 |
Wider availability of genetic testing, along with recognition that about 5% of cases of systemic amyloidosis are familial, has led to ever increasing numbers of new amyloidogenic and non-amyloidogenic mutations in these genes being identified. We have addressed the clinical and scientific need for a readily accessible repository of mutations and phenotypes in hereditary amyloidosis by creating the first online registry (www.amyloidosismutations.com) with the aim of collecting information on all known variants causing amyloidosis, including their reported clinical phenotype. Another objective was to introduce correct updated nomenclature for describing genetic variants. The current nomenclature used in practice is based on the processed gene product, therefore does not comply with the guidelines set by the Human Genome Variation Society (HGVS; www.hgvs.org/mutnomen) [den Dunnen and Antonarakis, 2000] and HUGO Gene Nomenclature Committee (HGNC). This has become especially important since the introduction of mass spectrometric analysis for fibril typing of amyloid deposits, where use of nomenclature based on the processed protein may result in mis-classification of newly identified amyloidogenic protein variants.
Registry design
The home page of the registry opens with a tabular list of ten buttons. The first eight give access to the disease gene of choice, one for reporting new mutations and the last button providing a link to an online discussion forum (Fig. 1).

Specific information on mutations identified in transthyretin (TTR; MIM# 176300), apolipoprotein AI (APOA1; MIM# 107680), apolipoprotein AII (APOA2; MIM# 107670), lysozyme (LYZ; MIM# 153450), fibrinogen α-chain (FGA; MIM# 134820), gelsolin (GSN; MIM# 137350) beta-2-microglobulin (B2M; MIM# 109700) and cystatin C (CST3; MIM# 604312) genes is provided. For each gene, the user can either open a graphical representation of whole or part of the cDNA sequence or a tabular list of mutations with the following information: change at the protein and nucleic acid levels, location of each variant, the type of alteration, the type and number of bases altered, a short description of the known clinical phenotype, ethnicity where the mutation was reported/is common and a link to the Medline citation, if relevant. The citation refers to the first author(s) who identified the mutation(s) either by publication or by personal submission. The distribution of mutations along the cDNA sequence is shown and the user can easily access each variant by simply clicking on the variant name. Hovering the cursor over a variant shows a brief summary of the mutation including the original citation. Clicking the variant name on the cDNA sequence takes the user to specific mutation in the tabular mutation list. The registry has a search facility which provides the user with a quick way to find and extract specific data. This home page has a submit button allowing for submission of novel mutations. There are also links to other web pages providing additional information about each gene including the Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/Omim), the genomic, cDNA and protein NCBI reference sequences.
Software and Search Engine
The registry was designed using Dreamweaver CS 5.5. The background scripting was done using HTML, JavaScript and PHP. Modification and additions are done locally and then transferred to live server using FTP. Mutation lists are stored on a MySQL database with various tables. The MySQL database is based on Linux platform and has its own control panel, which is accessible via web browser such as Internet Explorer. Any changes and additions of new information are done by the curators. The registry has built-in search engine optimization (SEO) and is searchable by most of the internet search engines.
The design of this registry was funded by Sixth European Union framework grant EURAMY (Systemic Amyloidosis in Europe). The registry is maintained by the UK National Amyloidosis Centre.
Updating Nomenclature
The nomenclature currently used by the amyloid community for describing mutations has not followed the general rule of starting with methionine as the translation initiator. As a consequence, almost all amyloidogenic variants described to date have amino acid numbering based on the processed gene product. Since this is not universally recognised and especially as it is not in keeping with current nomenclature recommendations, it has potential to cause serious errors. With the aim to move the amyloidosis gene nomenclature in accordance with current practice, we have designed this registry to include the description that complies with the HGNC and HGVS nomenclature guidelines: cDNA numbering begins at nucleotide 1, which is the A of the ATG translation initiation codon, protein sequence represents the primary translation product and thus includes the signal peptide. For convenience and to avoid confusion, description of protein variants includes both the ‘usual name’ and, in brackets, the nomenclature recommended by HGNC (http://www.genenames.org) and HGVS (http://www.hgvs.org/mutnomen); for example: TTR Val30Met (p.Val50Met); FGA Gly26Arg (p.Gly50Arg); GSN Asp187Asn (p.Asp214Asn). Thus far there is no report of intronic mutations associated with familial amyloidosis. All reference sequences for cDNA and proteins were obtained from the NCBI registry.
Mutation Information
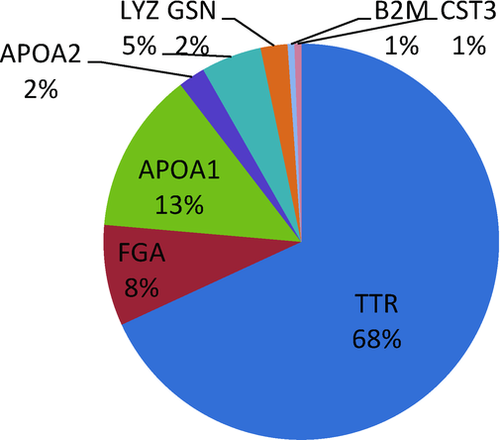
Currently the registry lists 182 variants of genes known to cause hereditary amyloidosis. TTR mutations account for 68%, APOA1 for 13%, FGA for 8%, LYZ for 5%, APOA2 for 2%, GSN for 2%, B2M for 1% and CST3 for 1% (Fig. 2).
The vast majority (91%) of reported variants are nonsynonymous point mutations, however frameshifts and deletions also occur. Nine percent of variants have been reported as non-pathogenic or of unknown clinical significance. The ethnic distribution of mutations is very broad and includes German, Portuguese, Swedish, Spanish, Italian, French, Polish, Russian, Japanese, Chinese, British, Irish, African-American, Indian, Finnish, Dutch and other populations.
A comprehensive search of the published literature enabled us to gather information about each mutation described to date. In addition, the registry includes 18 novel unpublished mutations (8 in TTR, 4 in APOA1 and 6 in FGA) of these 16 were identified in UK National Amyloidosis Centre and 2 independent submissions to the registry. The details of these mutations and their clinical phenotype are listed in Table 2.
| Gene | Name (Protein Variant) | Sequence Variant (mRNA) | Codon Change | Location | Reported Phenotype | Ethnic Group |
|---|---|---|---|---|---|---|
| TTR | Phe44Leu (p.Phe64Leu) | c.190T>C | TTT>CTT | Exon 2 | H | Caucasian |
| TTR | Thr49Ser (p.Thr69Ser) | c.206C>G | ACC>AGC | Exon 3 | PN | Indian |
| TTR | Glu54Leu (p.Glu74Leu) | c.220_221delinsTT | GAG>TTG | Exon 3 | H | Belgian |
| TTR | Glu54Gln (p.Glu74Gln) | c.220G>C | GAG>CAG | Exon 3 | H, PN | Romanian |
| TTR | Glu72Gly (p.Glu92Gly) | c.275A>G | GAA >GGA | Exon 3 | H | Caucasian |
| TTR | Ala81Val (p.Ala101Val) | c.302C>T | GCA>GTA | Exon 3 | H | Russian, Polish |
| TTR | His90Asp (p.His110Asp) | c.328C>G | CAT>GAT | Exon 3 | H | British |
| TTR | Ile107Phe (p.Ile127Phe) | c.379A>T | ATT>TTT | Exon 4 | AN, PN | British |
| APOA1 | Gly35Val (p.Gly59Val) | c.176G>T | GGC>GTC | Exon 3 | Unknown significance | Caucasian |
| APOA1 | Trp50Arg (p.Trp74Arg) | c.220T>A | TGG>AGG | Exon 4 | Renal impairment | Danish |
| APOA1 | Ala164Ser (p.Ala188Ser) | c.562G>T | GCC>TCC | Exon 4 | Unknown significance | Caucasian |
| APOA1 | Gln172Pro (p.Gln196Pro) | c.587_588delinsCC | CAG>CCC | Exon 4 | Likely amyloidogenic | Caucasian |
| FGA | Phe521Serfs*27 (p.Phe540Serfs*27) | c.1619_1622delTTGT | Frame shifting mutation | 5′ end of exon5 | Renal failure | Arab |
| FGA | Glu524Lys (p.Glu543Lys) | c.1627G>A | GAG>AAG | 5′ end of exon5 | Renal failure | Caucasian |
| FGA | Glu526Lys (p.Glu545Lys) | c.1633G>A | GAG>AAG | 5′ end of exon5 | Renal failure | Russian |
| FGA | Gly555Phe (p.Gly574Phe) | c.1720_1721delinsTT | GGT>TTT | 5′ end of exon5 | Renal failure | Norwegian |
| FGA | Gly519Arg (p.Gly538Arg) | c.1612G>A | GGA>AGA | 5′ end of exon5 | non-pathogenic/ unknown significance | British |
| FGA | Gly519Glu (p.Gly538Glu) | c.1613G>A | GGA>GAA | 5′ end of exon5 | non-pathogenic/ unknown significance | British |
Submission of new mutations and online discussion forum
Submission of new mutation to the registry is available via a button on the home page or from any of the genes. Selecting this button leads to an online submission form which consists of the following fields: author(s) name and email address, name and number of change at protein and cDNA level, location of mutation, phenotype and patient's ethnicity. On the form, next to each box, there is an example on how the data should be entered on the form (Fig. 3). Additionally, by ticking the relevant box(s) on this form authors would have to specify method used to detect amyloid deposit. A new mutation would be considered pathogenic if there would be sufficient clinical information provided by the author, histological evidence of amyloid deposition and appropriate amyloid fibril type was confirmed by IHC and/or LMD/MS. On submission, the form is directed to the curators who review the information and add it to the registry. This should permit faster and more efficient collection of appropriate information. All mutations submitted will be referenced to the submitting author and/or the appropriate Medline citation if available.

The registry also includes an online discussion forum which the curators hope will be a portal for discussion by and for the amyloidosis genetic community as well as a mechanism for anyone to communicate with the curators of the registry.
Discussion
Hereditary amyloidosis is an autosomal dominant disease with a variable penetrance. Most affected individuals are heterozygous although rare homozygous cases have been reported [Holmgren et al., 1992; Yoshinaga et al., 1994]. Hereditary amyloidosis can clinically present form early childhood upwards, but is usually a late-onset disease. Hereditary transthyretin (ATTR) amyloidosis is the commonest and has greatest genetic variability. More than 120 amyloidogenic TTR mutations have been described. ATTR amyloidosis usually presents with peripheral and autonomic neuropathy, but can also present as cardiac amyloidosis [Saraiva, 2002; Connors et al., 2003; Koike et al., 2009]. The TTR V122I variant is present in about 4% of the Afro-Caribbean population and is associated with late onset cardiac amyloidosis in an undetermined proportion of cases [Jacobson et al., 1997]. Hereditary non-neuropathic systemic amyloidosis [Ostertag, 1932] is associated with mutations in the APOA1 [Jones et al., 1991; Soutar et al., 1992; Booth et al., 1995; Persey et al., 1998; Hamidi Asl et al., 1999; Obici et al., 1999; de Sousa et al., 2000; Murphy et al., 2004; Rowczenio et al., 2011], APOA2 [Benson et al., 2001], LYZ [Pepys et al., 1993] and FGA [Benson et al., 1993; Uemichi et al., 1994; Hamidi Asl et al., 1997; Gillmore et al., 2009] genes. The clinical amyloidosis syndromes caused by the various mutations are remarkably diverse with respect to age of onset, mode of presentation, pattern of organ distribution, rate of progression, and prognosis. Renal involvement occurs with most mutations; however amyloid deposition can affect any or all of the major viscera. Apolipoprotein AI amyloidosis can occasionally present with neuropathy [Hazenberg et al., 2009; Nichols et al., 1990].
Whilst the most commonly identified variants are well documented with good information on the clinical phenotype and natural history, information on rarer variants including their amyloidogenic potential is difficult to obtain. Clinical significance of new mutations is often unclear due to lack of family history, reduced penetrance and other factors. Prior to the creation of this registry, there was no central repository containing detailed information. Small databases for individual gene mutations have been available but information on mutations in this disease generally was scattered. The idea to create an online registry of mutations in hereditary amyloidosis and associated clinical phenotypes was first proposed at the EU FP6 EURAMY meeting in 2007 and the UK National Amyloidosis Centre was put in charge of this project. First, we gathered information on mutations reported in each gene by comprehensive search of the published literature. In 2010, we centralized this data by establishing the registry and search tool, at www.amyloidosismutations.com, with the aim to make it easily accessible to the scientific and medical communities. Since its launch the website has been regularly updated with new variants identified from publications or those identified in our own centre. Since the start in 2010, a total of 47 new mutations (including the 18 mutations listed in Table 2), have been added to the registry.
Correct mutation nomenclature is essential. Compliance with the HGNC and HGVS nomenclature guidelines in the registry gives access to both “common” and correct gene nomenclature. This will allow researchers to have a uniform platform for efficient designing of tests and accurate reporting, and should ideally be used in all diagnostic laboratories. We hope this will prevent future confusion in describing genetic variants in this disease. The ability to submit new mutations online provides an efficient method for collecting and disseminating information. Currently, the registry contains 182 sequence variants in related genes; we hope that this number will increase in the future as we encourage clinicians and geneticist to collaborate in submission of novel variants to this registry. Routine use by the entire amyloidosis community will allow this resource to eventually help accurate documentation of genotype/phenotype correlations.
In summary, we report the first registry of mutations in hereditary amyloidosis as a free online resource for the clinical and scientific community for accessing information about and reporting new mutations and phenotypes in hereditary amyloidosis.
Acknowledgments
This work was supported by the EU FP6 EURAMY project; Work package 3.




