Mutations in Exon 1 Highlight the Role of MED12 in Uterine Leiomyomas
Contract grant sponsors: Academy of Finland (Academy Research Fellow grants 260370 and 265124); the Sigrid Jusélius Foundation; the Cancer Society of Finland; the Emil Aaltonen Foundation; the Orion-Farmos Research Foundation; the US Department of Health and Human Services National Institutes of Health (grants MH085320 and AR053100).
Communicated by Georgia Chenevix-Trench
ABSTRACT
Mediator regulates transcription by connecting gene-specific transcription factors to the RNA polymerase II initiation complex. We recently discovered by exome sequencing that specific exon 2 mutations in mediator complex subunit 12 (MED12) are extremely common in uterine leiomyomas. Subsequent screening studies have focused on this mutational hot spot, and mutations have been detected in uterine leiomyosarcomas, extrauterine leiomyomas and leiomyosarcomas, endometrial polyps, and colorectal cancers. All mutations have been missense changes or in-frame insertions/deletions. Here, we have analyzed 611 samples representing all above-mentioned tumor types for possible exon 1 mutations. Five mutations were observed, all of which were in-frame insertion/deletions in uterine leiomyomas. Transcriptome-wide expression data revealed that MED12 exon 1 and exon 2 mutations lead to the same unique global gene expression pattern with RAD51B being the most upregulated gene. Immunoprecipitation and kinase activity assays showed that both exon 1 and exon 2 mutations disrupt the interaction between MED12 and Cyclin C and CDK8/19 and abolish the mediator-associated CDK kinase activity. These results further emphasize the role of MED12 in uterine leiomyomas, show that exon 1 and exon 2 exert their tumorigenic effect in similar manner, and stress that exon 1 should be included in subsequent MED12 screenings.
Introduction
Uterine leiomyomas (fibroids) are the most common human tumors affecting nearly 70% of women by the age of 50 years [Day Baird et al., 2003]. They can have a major impact on women's health and quality of life by causing numerous symptoms such as abdominal pain, abnormal menstrual bleeding, pregnancy complications, and even infertility [Parker, 2007]. Uterine leiomyomas are the most common indication for hysterectomy, and therefore cause substantial costs for the health care system [Flynn et al., 2006]. Complex chromosomal rearrangements are frequent in these tumors, occasionally leading to tumor promoting changes such as overexpression of high mobility group AT-hook 2 (HMGA2) [Schoenmakers et al., 1995; Mehine et al., 2013]. While most uterine leiomyomas are sporadic, heterozygous germline mutations in the fumarate hydratase (FH) gene predispose individuals to cutaneous and uterine leiomyomas and renal cell cancer in the context of hereditary leiomyomatosis and renal cell cancer (HLRCC) syndrome (MIM #150800) [Tomlinson et al., 2002].
Recently, we utilized exome sequencing and identified somatic mediator complex subunit 12 (MED12; MIM #300188) mutations in as many as 70% (159/225) of uterine leiomyomas studied [Mäkinen et al., 2011a]. All mutations resided in exon 2 or in the preceding exon–intron boundary, and the majority of them affected a single codon encoding glycine 44 (69% of the observed mutations; 110/159). The original gene identification effort was based on leiomyomas collected from Caucasian women, and the finding has now been validated in other populations and ethnic groups [Mäkinen et al., 2011b; Je et al., 2012; Markowski et al., 2012a, 2012b; Matsubara et al., 2012; McGuire et al., 2012; Perot et al., 2012]. Of note, these subsequent mutation screenings have focused only on exon 2. The majority of observed mutations have been missense changes affecting specific mutational hot spots, but insertions and deletions (indels) leading to an in-frame transcript have also been found. According to cDNA sequencing, the mutant alleles are predominant, indicating that the mutations reside in the active X chromosome [Mäkinen et al., 2011a; Markowski et al., 2012a; McGuire et al., 2012; Perot et al., 2012]. The frequency of MED12 exon 2 mutations has been studied extensively also in other smooth muscle tumors, both benign and malignant, as well as in various mesenchymal and epithelial tumors. Mutations have been observed recurrently in uterine leiomyosarcomas, and with lower frequencies in extrauterine leiomyomas and colon carcinomas. Single mutations have been detected in endometrial polyps, smooth muscle tumors of uncertain malignant potential, and in extrauterine leiomyosarcomas [Cancer Genome Atlas Network, 2012; Je et al., 2012; Kämpjärvi et al., 2012; Markowski et al., 2012b; Matsubara et al., 2012; Perot et al., 2012; Ravegnini et al., 2012; de Graaff et al., 2013; Rieker et al., 2013; Schwetye et al., 2014].
MED12 is part of the mediator, which is a large, highly conserved protein complex that participates in the regulation of general as well as gene-specific transcription [reviewed in Taatjes, 2010]. MED12 forms the kinase/CDK8 module together with CDK8/CDK19, Cyclin C, and MED13 [Borggrefe et al., 2002]. Recent functional analyses have shown that exon 2 mutations in MED12 disrupt a MED12–Cyclin C interface in mediator, triggering loss of Cyclin C–CDK8/19 and thus mediator-associated kinase activity [Turunen et al., 2014]. Structurally, the Cyclin C-binding domain on MED12 resides within its N-terminal 100 amino acids, encoded largely by exons 1 and 2, indicating that leiomyoma-linked exon 2 mutations in MED12 disrupt its direct binding interface for Cyclin C [Turunen et al., 2014]. Notably, truncation of the first 20 amino acids of MED12, encoded solely by exon 1, significantly impaired Cyclin C binding, thus revealing that exon 1 must also encode important structural determinants of kinase module integrity.
The aim of this study was to analyze whether additional MED12 mutations can be found in exon 1, which at the transcript level resides right next to the exon 2 mutation hot spot. Altogether, 611 tumor samples representing all tumor types where MED12 exon 2 mutations have previously been observed were included in the study.
Materials and Methods
Six hundred and eleven MED12 exon 2 mutation-negative tumors were analyzed for possible MED12 exon 1 mutations. The sample series included uterine leiomyomas (conventional, various histopathological variants, and tumors from HLRCC patients), extrauterine leiomyomas, endometrial polyps, uterine leiomyosarcomas, other sarcomas, and colorectal cancer (CRC) samples. See Table 1 for more details on the sample series.
| Tumor type/histological subtype | n | FFPE/fresh frozen | Samples from | Reference |
|---|---|---|---|---|
| Leiomyomas from Caucasian patients | ||||
| Uterine leiomyoma | ||||
| Conventional | 73 | Fresh frozen | HUCH | [Mäkinen et al., 2011a] |
| Cellular | 49 | FFPE | PH | [Mäkinen et al., 2013] |
| Atypical | 15 | FFPE | PH | [Mäkinen et al., 2013] |
| Mitotically active | 15 | FFPE | PH | [Mäkinen et al., 2013] |
| From HLRCC patients | 34 | FFPE | PH | [Mäkinen et al., 2013] |
| Extrauterine leiomyoma | 39 | FFPE | CFCH/PH | [Kämpjärvi et al., 2012/Kiuru et al., 2002] |
| Leiomyomas from South African patients | ||||
| Uterine leiomyoma | 13 | Fresh frozen | UCT | [Mäkinen et al., 2011b] |
| Endometrial polyp | 55 | FFPE | PH | [Kämpjärvi et al., 2012] |
| Uterine leiomyosarcoma | ||||
| Early onset | 24 | FFPE | PH | [Ylisaukko-oja et al., 2006] |
| Unselected | 13 | FFPE | CFCH | [Kämpjärvi et al., 2012] |
| Sarcoma | ||||
| Soft tissue sarcoma | 79 | Fresh frozen | PH | [Kämpjärvi et al., 2012] |
| Bone sarcoma | 19 | Fresh frozen | PH | [Kämpjärvi et al., 2012] |
| Colorectal cancer (CRC) | ||||
| MSS | 83 | Fresh frozen | FCH | [Aaltonen et al., 1998, Salovaara et al., 2000] |
| MSI | 100 | Fresh frozen | FCH | [Aaltonen et al., 1998, Salovaara et al., 2000] |
| Tumor type | Sample ID | Ethnicity | Nucleotide changea | Amino acid change |
|---|---|---|---|---|
| Conventional uterine leiomyoma | M4m2 | Caucasian | c.76_91del16insG | p.P26_Q31delInsE |
| Conventional uterine leiomyoma | MY24m6 | Caucasian | c.82_99del18 | p.D28_E33del |
| Conventional uterine leiomyoma | MY32m13 | Caucasian | c.82_99del18 | p.D28_E33del |
| Conventional uterine leiomyoma | MY33m4 | Caucasian | c.80A>T; c.84_98del15 | p.Q27L; p.D28_K32del |
| Conventional uterine leiomyoma | FG166_1 | South African, mixed origin | c.77C>T; c.79_99del21 | p.P26L; p.Q27_E33del |
- a MED12 NM_005120.2 was used as a cDNA reference sequence.
- FFPE, formalin fixed paraffin embedded; HUCH, Helsinki University Central Hospital; PH, Department of Pathology at Helsinki University Central Hospital; CFCH, Central Finland Central Hospital; UCT, Department of Obstetrics and Gynecology at University of Cape Town; FCH, Finnish Central Hospitals; MSS, microsatellite stable; MSI, microsatellite unstable.
The study was approved by the Ethics Review Board of the Hospital District of Helsinki and Uusimaa (HUS), Helsinki, Finland, and the appropriate research permissions were obtained from local ethics committees. All samples were collected either after signed informed consent or after the approval by the director of the health care unit (anonymized samples).
MED12 (RefSeq accession number NG_012808.1) exon 1 mutation status was determined by direct Sanger sequencing. Genomic DNA from the fresh frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples was extracted as previously described [Kämpjärvi et al., 2012]. Total RNA was extracted with TRI Reagent® (Molecular Research Center Incorporated, Cincinnati, OH) according to manufacturer's instructions and cDNA conversion was performed with standard methods. Two primer pairs were used to amplify the genomic region of MED12 exon 1. Primers 5′ CCTCCGGAACGTTTCATAGAT 3′ (forward) and 5′ TTCGGGACTTTTGCTCTCAC 3′ (reverse) were used for the fresh frozen tissue samples, and primers 5′ CCCCTTTTCGGCTCCCTC 3′ (forward) and 5′ GTCAGTGCCTCCTCCTAGG 3′ (reverse) were used for FFPE tissue samples. Primer pair 5′ CTTCGGGATCTTGAGCTACG 3′ (forward) and 5′ GATCTTGGCAGGATTGAAGC 3′ (reverse) was used for cDNA sequencing. PCR conditions are available upon request. Sanger sequencing was performed on an ABI3730 Automatic DNA Sequencer (Applied Biosystems at Institute for Molecular Medicine Finland (FIMM) Genome and Technology Centre, Helsinki, Finland), and sequences were analyzed both manually and with a Mutation Surveyor Software (Softgenetics, State College, PA). MED12 NM_005120.2 was used as a cDNA reference sequence, where the A of the ATG translation initiation codon was used as +1 in nucleotide numbering. Observed MED12 mutations have been submitted to the locus-specific database (www.lovd.nl/MED12).
Gene expression profiling was performed with Affymetrix GeneChip Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA) using remapped Brainarray Custom CDF files (HuEx10stv2_Hs_ENSG, Version 17). All the arrays were hybridized and assessed for quality in the Biomedicum Functional Genomics Unit (FuGU, Helsinki, Finland) using the instructions provided by Affymetrix. Gene expression data were analyzed with Partek Genomic Suite™ v. 6.5 (Partek Incorporated, St. Louis, MO). All tumor and myometrium samples were quantile normalized by the Robust Multichip Average method and adjusted for probe sequence and GC content. Similarities between tumor samples were assessed using unsupervised hierarchical clustering analysis (cosine dissimilarity) with 1% most variable genes (n = 379), defined by the coefficient of variation calculated across all tumor samples.
Immunoprecipitations and kinase activity assays were performed as described in Turunen et al. (2014). In brief, site-directed mutagenesis (SDM) to generate MED12 exon 1 mutations reported in this study, p.P26_Q31delinsE, p.D28_E33del, and p.D28_K32del, was performed to pCDNA3.1 containing MED12 fragment (amino acids 1–593) followed by subcloning to the mammalian expression vector p3xFLAG–MED12. Primer sequences and conditions used in SDM are available upon request. pCDNA3.1–3xFLAG MED12 (WT or mutant derivative) plasmids were transfected to HEK293T cells, and FLAG-tagged products were immunoprecipitated with anti-FLAG M2 affinity gel (A2220; Sigma–Aldrich, St. Louis, MO) from cell lysates collected 48 hr later. FLAG-specific immunoprecipitates were washed and analyzed by Western blot with following antibodies: anti-FLAG M2 (F3165; Sigma–Aldrich), anti-MED23 (550429; BD Biosciences Pharmingen, San Diego, CA), anti-CDK19 (HPA007053; Sigma–Aldrich), anti-CDK8 (sc-1521; Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-MED4 produced by Kim et al. (2006), and anti-Cyclin C (558903; BD Biosciences Pharmingen). In vitro kinase assay was performed using [Y-32P]-ATP and a purified GST–CTD (C-terminal domain) substrate.
Results
Here, we have analyzed MED12 exon 1 in 611 tumors representing all tumor types where exon 2 mutations have previously been reported (Table 1). Altogether, five MED12 exon 1 mutations were observed (5/611, 0.8%), all in conventional uterine leiomyomas. All mutations were in-frame indels, four in uterine leiomyomas from patients of Caucasian origin (4/73; 5.5%) and one in a uterine leiomyoma from a South African patient with mixed ancestry (colored) (1/13; 7.7%; Table 1). Somatic origin of all these mutations was confirmed (Fig. 1A). Two leiomyomas derived from Caucasian patients harbored the same 18 nucleotide deletion c.82_99del18 (p.D28_E33del), the third was a deletion of 16 and insertion of one nucleotides c.76_91del16insG (p.P26_Q31delinsE), and the fourth was a deletion of 15 nucleotides c.84_98del15 (p.D28_K32del), accompanied by a point mutation c.80A>T (p.Q27L). Tumor tissue was available from all these four tumors and in all cases the mutant allele was predominantly expressed (Fig. 1A). One uterine leiomyoma from a South African patient harbored a deletion of 21 nucleotides; c.79_99del21 (p.Q27_E33del), accompanied by a point mutation c.77C>T (p.P26L). No RNA was available from this tumor. Detailed information on mutations is presented in Table 1. All MED12 exon 1 and exon 2 mutations in uterine leiomyomas reported in the original exome sequencing study and in our subsequent MED12 screenings are shown in Figure 1B. No MED12 exon 1 mutations were found in any other tumor type.
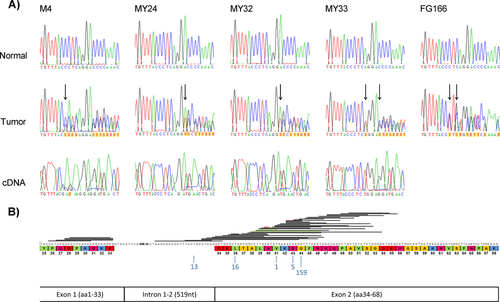
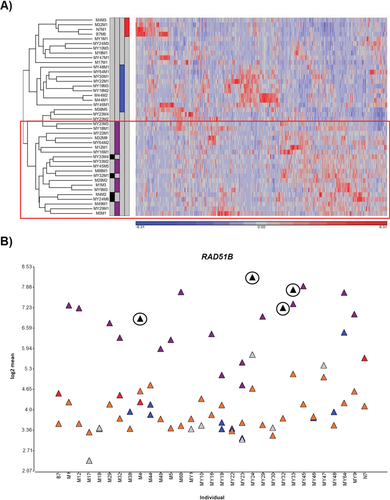
The functional impact of the observed mutations was first studied by transcriptome-wide gene expression profiling. RNA from four MED12 exon 1 mutation-positive tumors and their corresponding normal myometrium samples were available for the analyses. Data were analyzed together with the data from a previous study [Mehine et al., 2013], and the total sample series included 42 tumors and corresponding myometrium from 31 patients: four MED12 exon 1 mutation positive, 16 MED12 exon 2 mutation positive, four FH deficient, 10 HMGA1/HMGA2 overexpressing, and eight tumors lacking all these aberrations. Regardless of the mutations’ particular positions in the transcript, all MED12 mutation-positive samples displayed similar expression pattern, and the tumors clustered together in hierarchical cluster analysis (Fig. 2A). MED12 exon 1 mutation-positive tumors showed clear overexpression of RAD51B (Fig. 2B), which is the most significantly upregulated gene in MED12 exon 2 mutation-positive tumors [Mehine et al., 2013]. These results indicate that MED12 exon 1 and exon 2 mutations have the same and unique effect on global gene expression pattern.
To examine the effect of MED12 exon 1 mutations on mediator kinase module integrity, we performed immunoprecipitation and kinase activity assays from HEK293T cells expressing FLAG-tagged WT MED12 and three exon 1 and one exon 2 mutant derivatives. FLAG-specific immunoprecipitates were processed by Western blot analysis to monitor mediator subunit binding or in vitro kinase assay using a purified recombinant peptide substrate corresponding to the RNA polymerase II CTD. As expected, WT MED12 coprecipitated not only core mediator subunits, but also Cyclin C and CDK8/19 (Fig. 3B). Concordantly, WT MED12 immunoprecipitates also harbored robust CTD-directed kinase activity (Fig. 3C). In contrast, all MED12 exon 1 and exon 2 mutant derivatives coprecipitated significantly reduced levels of Cyclin C and CDK8/19, but not core mediator subunits (Fig. 3B) and bore little CTD-directed kinase activity (Fig. 3C). These results confirm that MED12 exon 1 mutations, like exon 2 mutations, uncouple Cyclin C-CDK8/19 from core mediator resulting in the loss of mediator-associated kinase activity.
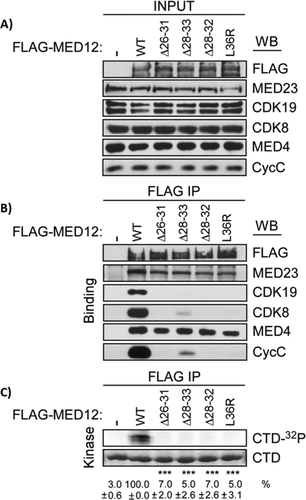
Discussion
Somatic MED12 exon 2 mutations are very common in conventional uterine leiomyomas, with frequencies of 52%–80% reported in various populations [Mäkinen et al., 2011a, 2011b; Je et al., 2012; Markowski et al., 2012b; Matsubara et al., 2012; McGuire et al., 2012; Perot et al., 2012]. Additional mutations in exon 1 detected in this study increase the proportion of MED12 mutation-positive lesions even further, thus highlighting the role of MED12 in the development of these tumors. Expression profiling and functional analyses indicate that exon 1 and exon 2 mutations have similar tumorigenic mechanisms, involving loss of mediator-associated CDK activity and RAD51B overexpression. This is in line with recent functional analyses showing that the first 100 amino acids of MED12 are central for its interaction with Cyclin C [Turunen et al., 2014]. Interestingly, the distribution of mutations is highly unbalanced with the vast majority affecting exon 2. The reason for this discrepancy remains thus far unexplained. Of note, no mutations in other exons of the gene have been observed in conventional uterine leiomyomas, neither in exome or whole-genome sequencing studies, nor in direct Sanger sequencing of the entire coding region of the gene [Mäkinen et al., 2011a; McGuire et al., 2012; Mehine et al., 2013]. Overall, the results of this study further highlight the role of MED12 in uterine leiomyomagenesis, and emphasize that exon 1 should be included in subsequent MED12 screenings.
Various treatment options are available for symptomatic uterine leiomyomas. These include hormone-mediated medical treatment, watchful waiting, and surgical interventions such as myomectomy, uterine artery embolization, and hysterectomy [Segars et al., 2014]. Medical treatment is preferred by most women, but current treatments reduce symptoms only temporarily and surgery remains a frequently utilized alternative. This means heavy economic burden for the health care systems as, for example, in the US alone, nearly 300,000 hysterectomies are performed each year due to leiomyomas [Jacoby et al., 2009]. The identification of specific MED12 exon 1 and exon 2 mutations in the vast majority of uterine leiomyomas promotes better understanding of the pathogenesis of these tumors, and provides an attractive target for drug development. Should MED12 mutation screening become a routine in a clinical setting, both exon 1 and exon 2 should be included in the analyses.
We have recently reported that various histopathological uterine leiomyoma variants harbor MED12 exon 2 mutations significantly less frequently than conventional leiomyomas (P = 2.93 × 10−8) [Mäkinen et al., 2013]. In this study, we found no exon 1 mutations in any of these rare leiomyoma subtypes. This further strengthens the hypothesis that the majority of these tumors arise through a distinct molecular mechanism than conventional uterine leiomyomas. Same applies to extrauterine and extrapelvic leiomyomas, where no exon 1 mutations were observed in this study and exon 2 mutations have previously been reported to be very rare [Kämpjärvi et al., 2012; Markowski et al., 2012a, 2012b; Matsubara et al., 2012; Ravegnini et al., 2012; de Graaff et al., 2013].
The only extrauterine cancer type where MED12 exon 2 mutations have been recurrently reported is colorectal cancer (CRC) [Cancer Genome Atlas Network, 2012; Je et al., 2012; Kämpjärvi et al., 2012]. While no exon 1 mutations were detected in this study, the analysis of CRC with next-generation sequencing has revealed two mutations affecting the last codon of exon 1, producing changes E33D and E33G [Forbes et al., 2010, Catalogue Of Somatic Mutations In Cancer; Cancer Genome Atlas Network, 2012]. These results suggest that MED12 exon 1 and exon 2 mutations may, albeit rarely, contribute also to the development of CRC.
Acknowledgments
We would like to thank Inga-Lill Svedberg, Mairi Kuris, Iina Vuoristo, Alison Ollikainen, Sini Nieminen, and Michael Parker for technical assistance.
Disclosure statement: The authors declare no conflicts of interest.




