Unraveling Cellular Phenotypes of Novel TorsinA/TOR1A Mutations
Contract grant sponsors: Hermann and Lilly Schilling Foundation; German Research foundation (LO 1555/3-2).
Communicated by Mark H. Paalman
ABSTRACT
A three-nucleotide (GAG) deletion (ΔE) in TorsinA (TOR1A) has been identified as the most common cause of dominantly inherited early-onset torsion dystonia (DYT1). TOR1A encodes a chaperone-like AAA+-protein localized in the endoplasmic reticulum. Currently, only three additional, likely mutations have been reported in single dystonia patients. Here, we report two new, putative TOR1A mutations (p.A14_P15del and p.E121K) that we examined functionally in comparison with wild-type (WT) protein and two known mutations (ΔE and p.R288Q). While inclusion formation is a characteristic feature for ΔE TOR1A, elevated levels of aggregates for other mutations were not observed when compared with WT TOR1A. WT and mutant TOR1A showed preferred degradation through the autophagy-lysosome pathway, which is most pronounced for p.A14_P15del, p.R288Q, and ΔE TOR1A. Notably, blocking of the autophagy pathway with bafilomycin resulted in a significant increase in inclusion formation in p.E121K TOR1A. In addition, all variants had an influence on protein stability. Although the p.A14_P15del mutation affects the proposed oligomerization domain of TOR1A, this mutation did not disturb the ability to dimerize. Our findings demonstrate functional changes for all four mutations on different levels. Thus, both diagnostic and research genetic screening of dystonia patients should not be limited to testing for the ∆E mutation.
Introduction
DYT1 dystonia is the most common form of isolated dystonia and is caused by autosomal dominantly inherited mutations in the TOR1A gene (MIM #605204). The vast majority of DYT1 cases are caused by the same in-frame three-base-pair deletion in Exon 5 of the TOR1A gene (c.904_906delGAG/c.907_909delGAG; p.302/p.303delE (ΔE)) [Ozelius et al., 1997; Klein et al., 1999]. At present, three additional mutations in TOR1A have been described in single patients (c.613T>A, p.F205I; c.863G>A, p.R288Q; c.966_983del, p.F323_Y328del) [Zirn et al., 2008; Leung et al., 2001; Calakos et al., 2010]. Moreover, an out-of-frame four-base-pair deletion was detected in a putatively healthy control (c.934_937del, p.R312Ffs*14) [Kabakci et al., 2004]. TOR1A encodes the 332 amino acid N-glycosylated protein TOR1A which is suggested to be a member of the AAA+ superfamily of chaperone proteins and among others functions in endoplasmatic reticulum-associated degradation [Ozelius et al., 1998; Neuwald et al., 1999; Hewett et al., 2000; Nery et al., 2011]. In primary cultures of neurons from wild-type (WT) and ΔE-knock-in mice, TOR1A protein was present in somata and dendrites [Koh et al., 2013]. Overexpression studies in cell models showed that WT TOR1A is targeted to the endoplasmic reticulum (ER) via its hydrophobic N-terminal signal sequence and is anchored to the luminal membrane of the ER, whereas the mutant ΔE forms membranous inclusions around the nucleus [Hewett et al., 2000; Kustedjo et al., 2000; Liu et al., 2003].
Nevertheless, the biology and function of TOR1A and how ΔE contributes to the development of DYT1 dystonia remain largely elusive. Moreover, diagnostic screening of dystonia patients is mainly limited to ΔE, although not all clinically typical cases can be ascribed to this mutation. For these reasons, we screened TOR1A in 162 ΔE-negative dystonia patients for other TOR1A mutations. By this, we detected two novel putative mutations (c.40_45delGCGCCG, p.A14_P15del; c.361G>A, p.E121K). To functionally characterize these novel and other reported putative mutations, we used different cell models and compared their properties with WT and ΔE TOR1A regarding subcellular localization, degradation pathways, protein stability, and dimerization.
Patients and Methods
Patients
The study was approved by the local ethics committee and all participants gave written informed consent. In our study, we included 162 patients with a positive family history for dystonia and/or onset <30 years who were diagnosed with focal (cervical dystonia, writer's cramp, blepharospasm, or musician's dystonia), segmental, or generalized dystonia. Patients positive for the ΔE mutation in TOR1A, or mutations in THAP1 or GNAL were not included. All patients were examined by a movement disorder specialist according to standard protocols. Secondary forms of dystonia were excluded by history and brain MRI. A cohort of 172 neurologically normal German controls [Kasten et al., 2013] was utilized to investigate the frequency of detected variants.
Mutational Analysis
To test for mutations, we sequenced all five coding exons including intron/exon boundaries as well as the TOR1A promoter region [Kaiser et al., 2010] of the TOR1A gene using a capillary sequencing machine (Life Technologies, Grand Island, NY, USA). Sequences were aligned to the genomic reference sequence (NG_008049.1) using the Mutation Surveyor software (SoftGenetics, State College, PA, USA). Coding DNA (cDNA) numbering is based on the coding reference sequence (NM_000113.2) and is given as “c.” followed by the position of the variant. This numbering refers to the A of the ATG translation initiation codon (Codon1) in the reference sequence as +1. For the in silico analysis of the investigated variants, five different prediction programs were used: MutationTaster (www.mutationtaster.org; [Schwarz et al., 2010]), SIFT (http://sift.jcvi.org/; [Kumar et al., 2009]), PROVEAN (http://provean.jcvi.org; [Choi et al., 2012]), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/; [Adzhubei et al., 2010]), and SNPs&GO (http://snps-and-go.biocomp.unibo.it/snps-and-go/; [Calabrese et al., 2009]). Accuracy of the prediction programs ranges from 65–91% [Calabrese et al., 2009; Kumar et al., 2009; Adzhubei et al., 2010; Schwarz et al., 2010; Choi et al., 2012]. The performance of several prediction methods has been evaluated in a recent study showing that SNPs&GO was one of the overall best performing methods [Thusberg et al., 2011].
Plasmid Construction and Mutagenesis
cDNA-encoding WT TOR1A was subcloned into the pcDNA3.1/myc-His vector (Life Technologies) and mutations (c.40_45delGCGCCG, c.361G>A, c.863G>A, c.904_906delGAG) were generated by site-directed mutagenesis (QuikChange; Stratagene, San Diego, CA, USA). For immunoprecipitation experiments, an HA-tag was introduced replacing the myc-tag for WT TOR1A. For the protein stability experiments, WT and mutant TOR1A cDNAs were subcloned into the pER4 vector and lentivirus was produced according to standard protocols [Park et al., 2008]. The TOR1A sequence was verified in all constructs by sequencing. In addition, WT and rs13300897:G>A TOR1A promoter plasmid [Kaiser et al., 2010] were used for luciferase reporter gene assays.
Cell Culture, Transfection, and Transduction
Human neuroblastoma SH-SY5Y cells (ATCC, Manassas, VA, USA) as well as HEK 293 FT cells (Life Technologies) were maintained in Dulbecco's modified Eagle's medium (PAA Laboratories (GE Healthcare), Little Chalfont, UK) supplemented with 10% fetal bovine serum (PAA Laboratories) and 1% penicillin/streptomycin (PAA Laboratories). For transient transfection of SH-SY5Y cells, 2 × 106 cells were transfected with 2 μg of cDNA using Nucleofector (Amaxa Cell Line Nucleofector kit V; Lonza, Basel, Switzerland). Cells were harvested 48 hr posttransfection. HEK 293 FT cells were transfected using the FuGene HD Transfection Reagent (Roche Diagnostics, Rotkreuz, Switzerland) and were harvested after 24 hr.
Luciferase Reporter Gene Assays
The WT minimal TOR1A promoter and the rs13300897:G>A variant were inserted into the pGL4.10 reporter gene plasmid (Promega, Fitchburg, WI, USA) to compare both native promoter activities. In addition, WT or mutant (p.H23P) THAP1 expression plasmids were used to investigate THAP1-mediated repression on both promoter constructs. HEK 293 FT cells were transiently transfected and luciferase activity was measured as described [Kaiser et al., 2010]. All experiments were carried out in at least three independent experiments and measured as triplicates, respectively.
Western Blotting
Protein concentration was determined using the Dc Protein Assay (BioRad, Hercules, CA, USA). For SDS PAGE, NuPAGE 4–12% Bis-Tris gels (Life technologies) were used. After electrophoresis, proteins were transferred to the nitrocellulose membrane (Protran (GE Healthcare), Little Chalfont, UK) and probed with primary antibodies raised against myc-Tag (Cell Signaling, Cambridge, UK) and β-tubulin (Sigma–Aldrich, St. Louis, MI, USA) as loading control.
Immunocytochemistry and Quantitative Analysis of the Intracellular Distribution of TOR1A
SH-SY5Y cells were fixed 48 hr after transfection in 4% paraformaldehyde. Myc-tagged TOR1A was visualized with a polyclonal anti-myc antibody (1:400; Cell Signaling Technology) and an Alexa 488 goat anti-rabbit antibody (Life Technologies). For ER staining, monoclonal antibody anti-protein disulfide isomerase (PDI, 1:1000; Abcam, Cambridge, MA, USA) and an Alexa 594 goat anti-mouse antibody (Life Technologies) were used. Cells were also stained with 1.0 μg/ml DAPI. The slides were analyzed with an LSM710 laser confocal microscope (Zeiss, Oberkochen, Germany) and images processed using Adobe Photoshop CS6 Extended (Adobe Systems Inc., San Jose, CA, USA).
Quantification of the localization of myc-tagged mutant and WT TOR1A was performed with unprocessed images of cells double-labeled for TOR1A and PDI, as a marker for the ER. By using the colocalization tool of the ImageJ (NIH) software, Pearson's coefficient was calculated (1 represents total colocalization and 0 no overlapping localization).
Treatment of Cells with Proteasome, Lysosome, and Autophagy Inhibitors
SH-SY5Y cells transfected with WT or mutant TOR1A were incubated for 24 hr at 37°C with the proteasome inhibitor MG132 (0.05 μM; Sigma–Aldrich), the lysosome inhibitors chloroquine (40 μM; Sigma–Aldrich) or NH4Cl (40 mM; Merck, Darmstadt, Germany), the autophagy inhibitor bafilomycin (3 nM; Sigma–Aldrich), or DMSO (Sigma–Aldrich) as vehicle. Next, cells were harvested by trypsinization and proteins were extracted using RIPA buffer (50 mM Tris–HCl, pH 7.6, 150 mM NaCl, 1% DOC, 1% NP-40, and 0.1% SDS). Finally, the lysates were centrifuged at 16,000g for 20 min at 4°C. The supernatants were used for Western blotting.
Assessment of Protein Stability Using a Doxycycline-Dependent TetOn System
For protein stability testing, SH-SY5Y cells were infected with two lentiviruses (MOI 3), one containing the Hygromycin-selectable regulatory plasmid transactivator tTA and the other one containing the response element TRE-Tight and the WT or mutant TOR1A coding sequence. Cells were treated for 3 hr in culture medium (DMEM, 10% FBS, 1% penicillin/streptomycin) supplemented with doxycycline (1 μg/ml). Next, cells were washed with PBS to remove doxycycline, further cultured and harvested after 1, 2, 3, and 4 days. SH-SY5Y cells infected with lentiviral tTA or with nonactivated Tet harboring both constructs served as controls. Samples were analyzed using SDS-PAGE and subsequent Western blotting.
Immunoprecipitation
HEK 293 FT cells plated in 10 cm dishes were cotransfected with HA-tagged TOR1A WT and myc-tagged WT or myc-tagged mutant p.A14_P15del. After 24 hr, cells were harvested and resuspended in 1 ml of lysis buffer (150 mM NaCl, 50 mM Tris–HCl, pH 7.6, 1% NP-40, 0.1% SDS, protease inhibitor cocktail [Roche Diagnostics]). Lysates were incubated on ice for 30 min and centrifuged at 13,000g for 20 min. The anti-HA antibody (5 μg) was bound to Dynabeads (Novax by Life Technologies) by incubation for 1 hr on a rotator. Then, the lysates, which were equalized for their protein concentration, were added to the washed antibody–Dynabeads complex and incubated for 1 hr on a rotator. After washing and elution, the supernatant was analyzed by Western blotting.
Formaldehyde Cross-Linking
HEK 293 FT cells were cotransfected with TOR1A HA-tagged and myc-tagged WT or with TOR1A HA-tagged WT and myc-tagged p.A14_P15del. After 24 hr, cells were pelleted and resuspended in 0.5% formaldehyde. Next, cells were incubated for 7 min at room temperature (RT) on a rotator and then pelleted at 1,800g at RT for 3 min. After removing the supernatant, the reaction was quenched with 0.5 ml 1.25 M glycine/PBS. Then, cells were spun again and washed once in 1.25 M glycine/PBS. Pellets were lysed in an adequate volume of RIPA buffer and protein concentration was determined using the Dc Protein Assay (BioRad). Samples were prepared for SDS-PAGE (without boiling of the samples) and subsequent Western blot analysis.
Statistical Analysis
One-way ANOVA with Dunnett's posttest was performed to evaluate the impact of inhibiting the protein degradation on the cells compared with the DMSO-control as well as for the comparison of Pearson's coefficients between WT and mutant TOR1A. One-way ANOVA with Sidak-Holm's posthoc test was used for evaluation of inclusion formation in non-treated SH-SY5Y cells overexpressing WT or mutant TOR1A. Inclusion formation in nontreated versus bafilomycin-treated SH-SY5Y cells overexpressing WT or mutant TOR1A, as well as between groups and treatments was examined by using a two-way ANOVA with posthoc Sidak-Holm's test. One-way ANOVA with posthoc Dunnett's test was performed for statistical analysis of protein stability testing. Differences between WT and p.A14_P15del mutant TOR1A in formaldehyde cross-linking experiments were examined by one-way ANOVA with Sidak-Holm's posttest. Significant differences between the measurements of all experiments were considered indicative for P values below 0.05.
Results
Patients and Mutation Screening
Clinical information on the 162 patients included in this study is given in Table 1. We identified three rare protein-changing sequence variants (1.9%, Table 2). First, a novel six base-pair deletion (c.40_45delGCGCCG, p.A14_P15del; http://databases.lovd.nl/shared/variants/0000036145) was found in one patient with cervical dystonia. This 76-year-old woman developed torticollis at the age of 31 years and some years later also a dystonic tremor of the head and hands. After a stereotactic operation in 1971, the symptoms improved. Since 2002, the patient has received local treatment with botulinum toxin A with a good response and the course of the disease remained stable. A follow-up neurological examination in 2011 at the age of 74 years, showed a torticollis and retrocollis. Further, we noticed impairment of repetitive movements of the left arm and leg, mild difficulties when writing, and mirror movements of both hands. With reference to the family history, the deceased mother had had similar symptoms (AAO > 75 years). No additional family members were available for segregation analyses. The detected deletion resides within exon 1 of the TOR1A gene and was not found among 172 neurologically normal German controls.
| Focal dystonia | |||||||
|---|---|---|---|---|---|---|---|
| All patients | Cervical dystonia | Blepharospasm | Writer's cramp | Musician's dystonia | Segmental dystonia | Generalized dystonia | |
| Number of patients | 162 | 27 | 16 | 7 | 34 | 37 | 41 |
| Male | 91 | 13 | 13 | 3 | 25 | 20 | 17 |
| Mean age (years) | 37.0 (±13.3) | 39.4 (±14.6) | 43.9 (±11.5) | 31.0 (±6.9) | 33.0 (±9.2) | 37.0 (±13.0) | 35.0 (±15.9) |
| AAO | 17.0 (±8.5) | 20.3 (±7.1) | 24.6 (±5.2) | 16.3 (±8.3) | 23.0 (±3.8) | 14.0 (±7.7) | 9.0 (±6.7) |
| Pos. family history | 45 | 7 | 15 | 3 | 6 | 3 | 11 |
| MAF among patients for p.A82A | 0.22 | 0.26 | 0.34 | 0.07 | 0.19 | 0.20 | 0.20 |
| MAF among patients for p.D216H | 0.15 | 0.15 | 0.03 | 0.14 | 0.13 | 0.18 | 0.18 |
- AAO, age at onset; MAF, minor allele frequency.
| Variants in TOR1A (NG_008049.1) | DNA changea | Amino acid change | Position | Interpretation | Frequency in our samples hom/het/WT (MAF) | Frequency in data baseb |
|---|---|---|---|---|---|---|
| New | c.40_45del | p.A14_P15del | Exon1 | mutation | 0/1/161 (0.003) | Not reported |
| Zirn et al. (2008) | c.863G>A | p.R288Q | Exon 5 | mutation | 0/1/161 (0.003) | Not reported |
| rs199535970 | c.361G>A | p.E121K | Exon2 | mutation? | 0/1/161 (0.003) | Not reported |
| rs13300897 | c.-104G>A | – | promoter | polymorphism | 8/49/102 (0.204) | 0.270 |
| rs114150156 | c.-52T>G | – | 5′UTR | polymorphism | 0/3/125 (0.012) | 0.010 |
| rs2296793 | c.264C>T | p.A82A | Exon 2 | polymorphism | 8/55/99 (0.219) | 0.280 |
| rs1801968 | c.646G>C | p.D216H | Exon 4 | polymorphism | 4/40/118 (0.149) | 0.150 |
- a Coding DNA numbering is based on the coding reference sequence (NM_000113.2) and is given as “c.” followed by the position of the base change. This numbering refers to the A of the ATG translation initiation codon (Codon1) in the reference sequence as +1.
- b Minor allele frequency (MAF) in the European population in the 1000Genomes database (http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/).
- hom, number of homozygous patients in our samples; het, number of heterozygous patients in our samples; WT, number of wild-type (WT) patients in our samples; MAF, minor allele frequency.
A second TOR1A variant (c.361G>A, p.E121K; http://databases.lovd.nl/shared/variants/0000036146) was identified in another patient (Table 2). The patient presented with segmental dystonia consisting of cervical dystonia and spasmodic dysphonia. At the time when we identified this variant, it was novel but is now reported in three individuals (3/13,006 = 0.0002) from the NHLBI Exome Sequencing Project (ESP; http://evs.gs.washington.edu/EVS/).
The third detected rare, protein-changing variant (c.863G>A, p.R288Q) was identified in one patient that has previously been reported [Zirn et al., 2008]. This only became obvious after having corresponded with the authors of the first description of this mutation.
Furthermore, several known SNPs were detected at comparable frequencies as in publically available databases (Table 2). This included a variant (rs13300897:G>A) within the predicted THAP1 binding site of the TOR1A promoter region. Although we did not detect any difference in the minor allele frequency between our patients (0.20) and our controls (0.19), we functionally tested for an effect on the strength of the TOR1A promoter. Using luciferase reporter gene assays, we neither detected any physiological effect on the TOR1A promoter activity nor on THAP1-mediated repression (Supp. Fig. S1).
To gain insight into possible effects of the protein-changing variants within TOR1A, an in silico analysis was performed using five different prediction programs (Table 3). Both programs that provided results for deletions indicated a disease-causing effect for the ΔE mutation, whereas only one of these tools categorized p.A14_P15del as pathogenic. For the substitution p.R288Q, two of five available programs predicted pathogenicity. The p.E121K variant was considered to be tolerated by all prediction tools.
| Variation | MutationTastera | SIFTb | Proveanc | Polyphen-2d | SNPs&GOe |
|---|---|---|---|---|---|
|
Disease causing | NA | Deleterious (score: −10.36) | NA | NA |
|
Polymorphism | NA | Deleterious (score: −3.47) | NA | NA |
|
Polymorphism | Tolerated (score: 1.00) | Neutral (score: 1.10) | Benign (score: 0.002) | Neutral |
|
Disease causing | Tolerated (score: 0.23) | Neutral (score: −1.61) | Possibly damaging (score: 0.955) | Neutral |
- a Based on NCBI37/Ensembl 69 [Schwarz et al., 2010].
- b Based on GRCh37/Ensembl63. The SIFT score ranges from 0 to 1 and declares an amino acid substitution damaging, if the SIFT score is ≤ 0.05 [Kumar et al., 2009].
- c Based on GRCh37/Ensembl66 and dbSNP database (Build 137). A threshold of -2.5 was used. Alterations with a score ≤-2.5 are predicted to be “deleterious” [Choi et al., 2012].
- d In Polyphen-2, a mutation is predicted to be “possibly damaging” if its Naïve Bayes probabilistic score is above 0.15. Mutations with a probabilistic score below 0.15 are predicted to be “benign” [Adzhubei et al., 2010].
- e SNPs&GO predicts “disease-related” or “neutral” mutation based on sequence and evolutionary information, and a defined functional GO score [Calabrese et al., 2009].
Subcellular Localization of WT and Mutant TOR1A and Inclusion Formation
To determine the effect of TOR1A mutations on the subcellular distribution of TOR1A, we performed double-labeling immunofluorescence confocal microscopic analysis in SH-SY5Y cells overexpressing myc-tagged TOR1A using antibodies against the myc-tag of TOR1A and the ER marker PDI (Fig. 1). WT TOR1A and the mutants p.A14_P15del, p.E121K, and p.R288Q showed a diffuse distribution in the cell with extensive colocalization with PDI (Fig. 1A, first four rows). In contrast, the mutant p.∆E hardly colocalized with anti-PDI staining but exhibited a perinuclear distribution with formation of membranous inclusions (Fig. 1A, bottom row). To quantify the degree of colocalized anti-PDI and TOR1A staining, we calculated the Pearson's coefficient (Fig. 1B) confirming an almost complete overlap for WT and all mutants but p.∆E TOR1A.
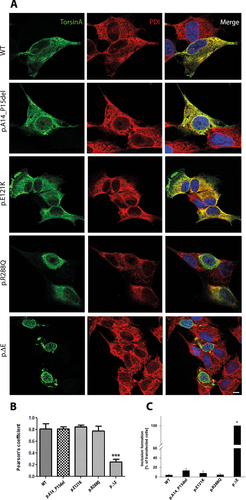
The formation of membranous inclusions is a well-known phenomenon for the p.∆E mutant [Hewett et al., 2000; Misbahuddin et al., 2005; Calakos et al., 2010]. In the present study, we used confocal immunofluorescence microscopy to quantify the number of inclusion-containing SH-SY5Y cells transfected with WT or the four different mutant forms of TOR1A. Under basal conditions, we confirmed inclusion formation in all cells transfected with p.∆E, whereas we observed inclusions in only 4–13% of the other transfected cells (Fig. 1C, Supp. Table S1).
Degradation Pathways of TOR1A
Previous inhibitor studies of TOR1A have shown that the degradation of TOR1A WT is mostly dependent on the autophagy-lysosome pathway. In addition, the degradation of mutant p.∆E also depends on selective proteasomal degradation [Giles et al., 2008; Gordon and Gonzalez-Alegre, 2008]. In light of this observation, we intended to explore the degradation pathways for the other potential mutations in comparison with WT and p.∆E (Fig. 2).
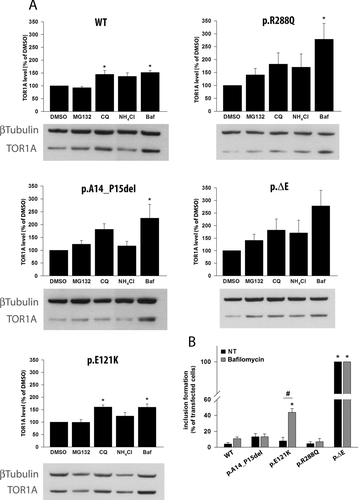
We examined the influence of proteasomal (MG132), lysosomal (chloroquine, NH4Cl), and autophagy (bafilomycin) inhibition on WT and mutant TOR1A (Fig. 2A). For WT, quantification of Western blot analysis showed a significant increase in TOR1A levels after chloroquine and bafilomycin treatment, whereas proteasome inhibition exhibited no effect which is in line with published data. Inhibition of the lysosome or the autophagosome leads to a 50% increase of the WT TOR1A protein. The p.∆E levels increased after treatment with all different inhibitors and were most pronounced after inhibition of the autophagy pathway (Fig. 2A, lower right, threefold increase). Similarly, p.A14_P15del and p.R288Q mutant TOR1A levels increased by about 2.5-fold after bafilomycin treatment, about twofold after chloroquine treatment, and only about 50% after blocking of the proteasome. The p.E121K mutant behaved like WT TOR1A after blocking the different degradation pathways (Fig. 2A).
Since bafilomycin had the strongest effect on TOR1A degradation in all TOR1A forms, we next tested whether inhibition of the autophagosome has an effect on inclusion formation. Bafilomycin treatment of p.E121K TOR1A leads to a significant increase of cells with inclusions (44% vs. 8%; Fig. 2B). Examples of the formation of inclusions in SH-SY5Y cells after bafilomycin treatment are shown in Supp. Figure S2. Information on cell counts and inclusion formation is provided in the Supp. Table S1.
Protein Stability of TOR1A Using the TetOn System
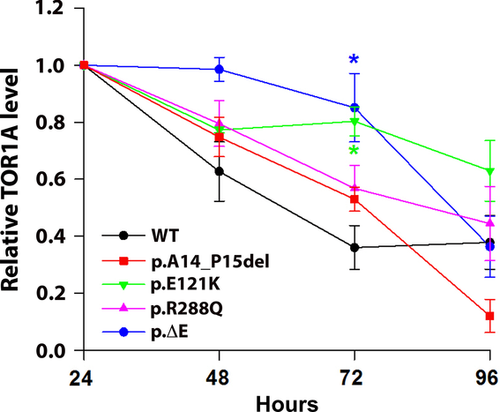
Next, we used a TetOn system with doxycycline donation to activate TOR1A protein production in order to investigate possible effects of the different mutations on TOR1A stability (Fig. 3). Following 3 hr-treatment with doxycycline, protein levels were higher for all variants after 72 hr compared with WT (significant for p.E121K and ΔE). On day 4, protein levels of p.A14_P15del decreased to 20%, whereas WT, p.R288Q, and ΔE remained at around 40% of the initial protein amount. The most stable TOR1A variant after 4 days is p.E121K at around 60% of the initial protein levels.
TOR1A p.A14_P15del Forms Oligomers
Coimmunoprecipitation studies showed that the N-terminal region of TOR1A is important for the ability of TOR1A to form oligomeric complexes [Giles et al., 2008]. Since our novel mutation p.A14_P15del is located in the N-terminal region of TOR1A, we performed coimmunoprecipitation experiments to investigate whether the mutated TOR1A was still able to bind WT TOR1A (Fig. 4A). These experiments showed that, despite the deletion of two amino acids, the ability of the mutant to bind WT TOR1A was not impaired.

Previously published sequence analysis suggested a shared hexameric structure of AAA+ family members [Neuwald et al., 1999]. This prompted us to further study the ability of our mutant p.A14_P15del to dimerize. We used formaldehyde cross-linking experiments, which confirmed that p.A14_P15del TOR1A formed dimers similar to WT TOR1A (Fig. 4B). Our data even suggest that mutant TOR1A protein favors dimerization, as the band at 74 kDa for the mutant was twice as strong as for WT (Fig. 4C).
Discussion
In this study, we identified a novel in-frame six-base-pair deletion (c.40_45delGCGCCG; p.A14_P15del) and two missense variants (c.361G>T, p.E121K; c.863G>A, p.R288Q) in the TOR1A gene in one patient each among 162 cases with isolated dystonia. To date, there are only four variants reported in TOR1A linked to isolated dystonia (p.F205I [Calakos et al., 2010], p.R288Q [Zirn et al., 2008], p.∆E [Ozelius et al., 1997], and p.F323_Y328del [Leung et al., 2001]). Numerous studies have shown that the p.∆E mutation, which is the most common cause of DYT1 dystonia, leads to a mislocalization of the mutant TOR1A from the ER to the nuclear envelope [Gonzalez-Alegre and Paulson, 2004; Goodchild and Dauer, 2004; Goodchild et al., 2005; Misbahuddin et al., 2005; Giles et al., 2008; Gordon and Gonzalez-Alegre, 2008]. The three mutations detected among our 162 patients were all mainly localized in the ER like overexpressed WT TOR1A and not within the nuclear envelope like p.∆E.
In addition, we selectively inhibited different degradation pathways to assess their role in the clearance of WT and the different mutant TOR1A proteins. Our results confirm previous studies, which have shown that WT TOR1A is predominantly degraded by the autophagy-lysosome pathway [Giles et al., 2008; Gordon and Gonzalez-Alegre, 2008]. Also TOR1A variants are degraded mainly by the autophagy pathway. Blocking of the autophagosome by bafilomycin lead to a moderate but significant increase of about 50% for WT and p.E121K TOR1A levels, whereas the effect was more pronounced for p.A14_P15del, p.R288Q, and p.∆E. Thus, the importance of the different degradation pathways was similar for p.A14_P15del and p.R288Q resembling the pattern for the established p.∆E mutation, whereas p.E121K behaved like the WT protein.
As a next readout in our mutants, we investigated the formation of membranous inclusions which is a well-described phenomenon caused by overexpression of p.∆E but not WT TOR1A [Gonzalez-Alegre and Paulson, 2004; Misbahuddin et al., 2005]. We confirmed the inclusion formation for p.∆E TOR1A. In contrast, WT and all other mutants formed inclusions in only 4–13% of the transfected cells under basal conditions. As inhibition of autophagy showed a strong effect for p.A14_P15del, p.R288Q, and p.∆E, we questioned whether there is a link to the formation of inclusions. Interestingly, bafilomycin treatment resulted in significantly increased formation of inclusions for the p.E121K variant, whereas the other mutants and WT were only slightly affected. Therefore, inhibition of autophagy is not generally linked to inclusion formation but shows a strong effect for the p.E121K mutation on aggregation.
An important aspect related to degradation pathways is the overall protein stability of WT and mutant TOR1A. To compare protein stability among different TOR1A forms, we used a doxycycline-dependent TetOn system to regulate the transcription of the TOR1A constructs. Examination of protein levels indicated a higher protein stability for all mutants three days postdoxycycline treatment compared with WT, which was most pronounced for p.E121K and ΔE. After 4 days, the relative protein levels changed considerably: Compared with WT, only p.E121K appeared to be more stable, whereas the protein levels of p.R288Q and ΔE reached comparable levels, and p.A14_P15del seemed to be even less stable. A previous study using pulse-chase labeling revealed that WT TOR1A is more stable than p.∆E mutant protein [Giles et al., 2008]. The differences between our results and the previously reported data may be ascribed to the different methods used. In the previous study, SH-SY5Y cells transiently transfected with TOR1A were employed. Thus, these cells had been growing for at least 24 hr in the presence of overexpressed TOR1A prior to radioactive pulse-labeling. This may have had an effect on the degradation machinery, which was already strongly activated in order to degrade the high amount of protein load. In contrast, our experiments were based on a short activation of TOR1A transcription for 3 hr in cells that were stably transduced, which results in a lower amount of protein expression and cellular stress.
Given that TOR1A belongs to the family of AAA+ proteins, its structure and ability to form oligomers is of importance for its function [Ozelius et al., 1997]. Our results suggest an enhanced dimerization of the p.A14_P15del mutant compared with WT TOR1A. A previous study showed that the N-terminal region of TOR1A is necessary for its ability to form oligomers [Giles et al., 2008]. However, although our novel mutation is located within the N-terminus of TOR1A, the deletion of two amino acids is insufficient to disturb this interaction.
In conclusion, we identified a considerably high frequency of 2% of rare protein-changing sequence variants in the TOR1A gene among patients with isolated dystonia. Detailed functional characterization of these variants points toward pathogenic changes of these altered TOR1A bears some analogy to p.∆E TOR1A. Thus, these variants may be considered as possible disease-causing mutations. The observed changes include an increased activation of the autophagy-lysosomal degradation pathway in p.A14_P15del and p.R288Q comparable to the one observed in p.∆E TOR1A. Overexpression of p.E121K resulted in increased inclusion formation upon inhibition of the autophagy-lysosomal degradation pathway, with inclusion formation being a well-known phenomenon for overexpressed p.∆E TOR1A. Moreover, all of the variants seem to have an influence on protein stability.
Additional experiments are needed to further elucidate the exact disease-causing mechanism(s) of these TOR1A mutants, as the number of observations and functional tests presented here is limited. However, it became obvious that TOR1A mutations other than the p.∆E mutation may be of pathophysiological relevance for isolated dystonia. Thus, both diagnostic and research genetic testing of dystonia patients should not be limited to screening for p.∆E. As demonstrated in our relatively limited patient sample, additional potentially pathogenic mutations can be detected at a considerable frequency even in smaller cohorts.
Acknowledgement
The project was funded by the Hermann and Lilly Schilling Foundation and the German Research foundation (LO 1555/3-2).
Disclosure statement: The authors declare no conflict of interest.




