Hypomorphic NOTCH3 Alleles Do Not Cause CADASIL in Humans
Contract grant sponsor: Brain Foundation of The Netherlands(F2009[1]-25).
Communicated by Jacques S. Beckmann
ABSTRACT
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is caused by stereotyped missense mutations in NOTCH3. Whether these mutations lead to the CADASIL phenotype via a neomorphic effect, or rather by a hypomorphic effect, is subject of debate. Here, we report two novel NOTCH3 mutations, both leading to a premature stop codon with predicted loss of NOTCH3 function. The first mutation, c.307C>T, p.Arg103*, was detected in two brothers aged 50 and 55 years, with a brain MRI and skin biopsy incompatible with CADASIL. The other mutation was found in a 40-year-old CADASIL patient compound heterozygous for a pathogenic NOTCH3 mutation (c.2129A>G, p.Tyr710Cys) and an intragenic frameshift deletion. The deletion was inherited from his father, who did not have the skin biopsy abnormalities seen in CADASIL patients. These individuals with rare NOTCH3 mutations indicate that hypomorphic NOTCH3 alleles do not cause CADASIL.
CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) is a monogenic disorder caused by mutations in the NOTCH3 gene (MIM #600276) [Joutel et al., 1996]. CADASIL patients typically present with stroke and cognitive decline at a mean age of 45–50 years. Migraine with aura and mood disturbances occurs in up to 40% of patients. Disease severity and age of onset is variable, also within families, and the clinical presentation can be atypical. The invariable and preclinical presence of a recognizable pattern of white matter hyperintensities (WMH) on brain MRI plays a key role in clinical diagnosis, seen in all individuals with a CADASIL-causing NOTCH3 mutation from at least 35 years of age. Next to increasingly extensive WMH, subcortical infarcts are typically seen in symptomatic patients, whereas cortical and cerebellar lesions are rare [Chabriat et al., 2009]. The clinical diagnosis is confirmed by NOTCH3-sequencing analysis, with the detection of a typical cysteine-altering mutation [Joutel et al., 1997]. If the clinical or molecular results are inconclusive, immunohistochemistry and electron microscopy (EM) can be performed on a skin biopsy, to determine the presence of pathognomonic vessel wall abnormalities. These consist of positive NOTCH3 staining, electron microscopic deposits of granular osmiophilic material (GOM), and vascular smooth muscle cell (VSMC) degeneration [Joutel et al., 2001; Tikka et al., 2009]. Next to a compatible clinical and family history, the trias of brain MRI, molecular NOTCH3 analysis, and, if necessary, skin biopsy usually allows for conclusive confirmation or rejection of CADASIL diagnosis.
NOTCH3 encodes the NOTCH3 protein, a transmembrane protein that, in adults, is predominantly expressed in the vasculature by VSMC and pericytes [Joutel et al., 2000]. The ectodomain of NOTCH3 (NOTCH3ect) consists of 34 epidermal growth factor-like repeat (EGFR) domains, which each contain exactly six highly conserved cysteine residues. NOTCH3 mutations in CADASIL are located in the exons encoding the NOTCH3ect (exons 2–23) [Chabriat et al., 2009], and invariably lead to an uneven number of cysteine residues, typically five or seven, in the corresponding EGFR. This leaves one cysteine unpaired and has been shown to lead to increased multimerization of the mutated protein [Duering et al., 2011]. In more than 95% of cases, the cysteine amino acid change is caused by a missense mutation [Chabriat et al., 2009]. Despite the stereotyped nature of NOTCH3 mutations in CADASIL, it is still unknown whether the primary determinant of the CADASIL phenotype is a hypomorphic or rather a neomorphic effect. The most adhered to hypothesis is that there is a neomorphic effect, with toxic accumulation of the mutated protein at the VSMC membrane [Joutel et al., 2000]. However, in a recent study, the loss-of-function hypothesis is revived, as a reduced NOTCH3 signaling was found to be associated with CADASIL pathophysiology in vitro and in vivo [Arboleda-Velasquez et al., 2011].
Here, we describe individuals from two families with NOTCH3 mutations that are predicted to result in a premature stop codon. We combined clinical, molecular, radiological, and pathological investigations to determine whether patients with these hypomorphic mutations have a CADASIL phenotype.
The index patient of family 1, a 55-year-old Caucasian male (index, II-2, Fig. 1A), was referred to our multidisciplinary CADASIL out-patient clinic, because a novel NOTCH3 variant with unknown pathogenicity had been detected in a laboratory elsewhere. The variant, a NOTCH3 nonsense mutation in exon 3 (c.307C>T, p.Arg103*) (NM_000435.2), was confirmed in the Leiden Laboratory for Diagnostic Genome Analysis in genomic DNA and in mRNA isolated from blood (Fig. 1B) (www.lovd.nl/NOTCH3). The case history reported a polyneuropathy, migraine with aura, and ischemic strokes at the age of 50 and 52 years. Neurological examination revealed mild motor weakness of the left hallux, absence of vibration sense in feet, difficulty in fine motor tasks of the left hand, and a bilateral action tremor. Mini-mental state examination (MMSE) was normal (30/30). Brain MRI (axial FLAIR and T2* gradient echo, sagittal T1 and coronal FLAIR, and T2 weighted images) was reviewed by a neuroradiologist specialized in CADASIL and showed three old infarctions, respectively, in the right and left occipital cortex and in the left parietal cortex, consistent with large vessel stroke. Furthermore, a few small circumscript WMH were seen. There were no lacunar infarctions and no WMH in a distribution or extent consistent with CADASIL (Fig. 1C). A skin biopsy was then taken from the inner side of the forearm, which showed a normal vessel wall structure, negative NOTCH3 staining (Novus Biologicals, Littleton, CO; dilution 1:2000) and no electron microscopic GOM deposits or VSMC degeneration (Supp. Fig. S1 A and B). Family history was negative for stroke and dementia. The index has three siblings, all older than 45 years of age. They all have a history of migraine but no other neurological symptoms. His mother (I-2) was tested negative for the NOTCH3 mutation and his father (I-1) is deceased, but had no medical history of stroke or dementia. His 50-year-old brother (II-3) had received genetic counseling elsewhere and had opted for predictive NOTCH3 mutation testing, showing the presence of the familial NOTCH3 nonsense mutation. This brother was then also referred to our CADASIL clinic, where neurological examination and brain MRI (axial T1, T2, FLAIR weighted images) were found to be completely normal (Fig. 1D).

In the second family, the index patient is a 40-year-old male (index, II-1; Fig. 2A), with a medical history of migraine with aura, bipolar disorder, and sarcoidosis. There was no history of (transient) neurological deficits. Neurological examination was normal, his MMSE was 28/30. Brain MRI (axial T1, T2, proton density, and sagittal FLAIR weighted images) had been made at a hospital elsewhere and showed partly confluent WMH in a pattern consistent with CADASIL, including the anterior temporal lobes (Supp. Fig. S2). To confirm the clinical diagnosis, blood was sent to the Leiden Laboratory for Diagnostic Genome Analysis for NOTCH3 mutation analysis. Sequence analysis was performed of exons 2–23, including intron/exon boundaries. A characteristic CADASIL causing NOTCH3 mutation (c.2129A>G, p.Tyr710Cys) was detected in exon 13 (Fig. 2B), which appeared to be homozygous. To exclude the possibility of a NOTCH3 deletion as the reason for the absence of the wild-type allele, multiplex ligation-dependent probe amplification (MLPA) analysis was performed using a homemade MLPA kit containing probes for all coding exons (2–33) of NOTCH3. This revealed the presence of a large intragenic heterozygous frameshift deletion of exons 3–16 (Fig. 2C). This patient is, therefore, compound heterozygous for a typical CADASIL-causing missense mutation and a large intragenic NOTCH3 deletion. Although the clinical diagnosis of CADASIL was confirmed by the detection of the NOTCH3 missense mutation, a family study was performed to determine the pathogenicity of the intragenic NOTCH3 deletion. NOTCH3 mutation analysis of the patient's parents showed that the p.Tyr710Cys mutation was inherited from his mother (I-2) and the NOTCH3 deletion was inherited from his father (I-1). A comprehensive medical assessment of the parents was complicated by the fact that they live in the former Dutch colonies. The medical history of the mother, however, was notable for a brain scan made around the age of 40 years after transient functional deficit of the arm. This scan was no longer available and she did not consent to further medical investigations. The 65-year-old father ostensibly has a history of psychiatric problems, but he has no history of stroke or dementia. He did not consent to brain imaging, but he did agree to have a skin biopsy taken, which was sent to us for pathological analysis and fibroblast culture. NOTCH3 staining of skin vasculature was negative and EM showed no GOM, VSMC degeneration, or other abnormalities (Supp. Fig. S1 C and D). To further determine the breakpoints of the NOTCH3 deletion, we performed NOTCH3 RT-PCR on RNA derived from the father's fibroblasts, using an exon 2 forward and an exon 18 reverse primer. Sequencing of the obtained RT-PCR product showed that exons 3–16 are completely missing as a result of the deletion (Fig. 2D). This is predicted to lead to a disruption of the NOTCH3 reading frame and a premature stop codon in exon 17 (r.198_2566del, p.Cys67ProfsX34) (www.lovd.nl/NOTCH3).
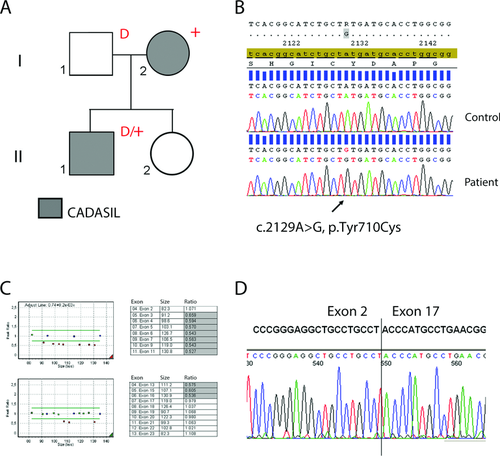
The novel NOTCH3 mutations detected in the two families described above are a nonsense mutation and an intragenic frameshift deletion, respectively. Both mutations are predicted to lead to a truncation at the protein's N-terminus, which then lacks essential domains required for NOTCH3 processing and signaling, including the transmembrane and intracellular domains. Therefore, these mutations must result in hypomorphic alleles, that is, alleles that have reduced gene activity. One individual with the nonsense mutation (c.307C>T, p.Arg103*; family 1, II-2) has a history of stroke, but his brain imaging is inconsistent with CADASIL and, moreover, his skin biopsy was negative for CADASIL vessel wall pathology. His brother (II-3), with the same nonsense mutation, was healthy at the age of 50 years and had a normal brain MRI. Together, these findings indicate that the NOTCH3 nonsense mutation is a coincidental finding and does not cause CADASIL. In family 2, the father of the index (I-1) has a NOTCH3 deletion of exon 3 through 16 on one allele. He allegedly has a history of psychiatric problems, but at the age of 65 years he has no medical history of stroke or dementia. Although a brain MRI would have contributed to a confirmation or exclusion of the diagnosis CADASIL, a skin biopsy was made that did not show any of the abnormalities consistently seen in CADASIL patients. As both the NOTCH3 nonsense mutation and the intragenic NOTCH3 deletion do not lead to a CADASIL phenotype, we conclude that hypomorphic NOTCH3 alleles do not cause CADASIL. This conclusion is further strengthened by the fact that frameshift and nonsense NOTCH3 variants have been reported previously in public databases and in an exome resequencing study on colorectal cancer patients (http://www.ncbi.nlm.nih.gov/snp/) [Boomsma et al., 2013; Smith et al., 2013]. Whole NOTCH3 gene deletions have also been reported in humans, but only in the context of contiguous gene deletions in patients with multiple congenital anomalies (http://decipher.sanger.ac.uk). In contrast to our findings, small out-of-frame NOTCH3 deletions, resulting in hypomorphic alleles, have been previously reported to be associated with CADASIL [Dotti et al., 2004; Weiming et al., 2013]. However, in these reports, the association of the hypomorphic allele with CADASIL is questionable; as the clinical diagnosis was not confirmed by skin biopsy, segregation analysis was not possible and in one study mutation analysis of the NOTCH3 gene was incomplete. The few small NOTCH3 deletions or insertions that have been proven to be the cause of CADASIL are all in-frame and typically affect cysteine residue number, or in one case, cysteine spacing [Dichgans et al., 2000, 2001; Mazzei et al., 2004, 2008; Tikka et al., 2009]. The fact that only cysteine-altering mutations have been found to be unequivocally associated with CADASIL, strongly supports the concept that cysteine-altering NOTCH3 mutations are a conditio sine qua non for CADASIL. In the clinical setting, mutations detected in NOTCH3 should therefore be carefully evaluated for their effect on cysteine residues in the NOTCH3 protein. If the mutation, be it missense or otherwise, does not affect the number of cysteines in an EGFR domain, then this mutation should be interpreted with great caution and CADASIL diagnosis should only be confirmed based on additional investigations, such as a skin biopsy. Also, when there is an apparent homozygous mutation, the presence of a deletion on one of the alleles should be excluded. An experienced team is important for the correct interpretation of atypical NOTCH3 mutations, to prevent an erroneous diagnosis of CADASIL. Such a team, in our opinion, should consist at least of a clinical geneticist, clinical laboratory geneticist, neurologist, genetic psychologist, (neuro)pathologist, and neuroradiologist.
Notably, we report herein the first CADASIL patient who is compound heterozygous for a typical cysteine-altering NOTCH3 mutation on one allele and a large intragenic NOTCH3 deletion on the other allele. Interestingly, this patient has a phenotype within the normal CADASIL spectrum.
In conclusion, our findings indicate that hypomorphic NOTCH3 alleles do not cause CADASIL in humans, which has important implications for diagnostic interpretation of NOTCH3 mutations. Furthermore, this can refine the focus of future studies on the effect of CADASIL-causing mutations on the NOTCH3 protein and the development of rational therapeutic approaches.
Acknowledgments
We thank Ingrid Hegeman for skin biopsy processing. In the former Dutch colonies, Denie Saimo-Abas is acknowledged for referral of patients, and Ester Lai-al-Fat for taking skin biopsies. From the LUMC Laboratory for Diagnostic Genome Analysis, we thank Merlijn van Nieuwenhuizen, Kirsten Heijboer, and Dave van Heusden for technical assistance.
Disclosure statement: The authors declare no conflict of interest.




