Coffin–Siris Syndrome and the BAF Complex: Genotype–Phenotype Study in 63 Patients
Communicated by Maria Rita Passos-Bueno
ABSTRACT
De novo germline variants in several components of the SWI/SNF-like BAF complex can cause Coffin–Siris syndrome (CSS), Nicolaides–Baraitser syndrome (NCBRS), and nonsyndromic intellectual disability. We screened 63 patients with a clinical diagnosis of CSS for these genes (ARID1A, ARID1B, SMARCA2, SMARCA4, SMARCB1, and SMARCE1) and identified pathogenic variants in 45 (71%) patients. We found a high proportion of variants in ARID1B (68%). All four pathogenic variants in ARID1A appeared to be mosaic. By using all variants from the Exome Variant Server as test data, we were able to classify variants in ARID1A, ARID1B, and SMARCB1 reliably as being pathogenic or nonpathogenic. For SMARCA2, SMARCA4, and SMARCE1 several variants in the EVS remained unclassified, underlining the importance of parental testing. We have entered all variant and clinical information in LOVD-powered databases to facilitate further genotype–phenotype correlations, as these will become increasingly important because of the uptake of targeted and untargeted next generation sequencing in diagnostics. The emerging phenotype–genotype correlation is that SMARCB1 patients have the most marked physical phenotype and severe cognitive and growth delay. The variability in phenotype seems most marked in ARID1A and ARID1B patients. Distal limbs anomalies are most marked in ARID1A patients and least in SMARCB1 patients. Numbers are small however, and larger series are needed to confirm this correlation.
Introduction
The April 2012 issue of Nature Genetics contained three articles linking variants in BRG1- and BRM-associated factor (BAF) complex components to intellectual disability syndromes. Variants in several components of the BAF complex were identified in patients with Coffin–Siris syndrome (CSS) [Santen et al., 2012a; Tsurusaki et al., 2012], and variants in SMARCA2 (MIM #600014) were linked to Nicolaides–Baraitser syndrome (NCBRS) [Van Houdt et al., 2012]. In addition, variants in ARID1B (MIM #614556) have been linked to nonsyndromic intellectual disability [Hoyer et al., 2012]. Although CSS and NCBRS are different clinical entities, there is some overlap, as both syndromes feature intellectual disability, hair abnormalities (hypertrichosis and sparse scalp hair), and digital abnormalities (short fifth fingers, prominent interphalangeal joints, prominent distal phalanges).
The BAF complex, one of the ATP-dependent chromatin remodeling complexes, mediates the opening and closing of chromatin [Hargreaves and Crabtree, 2011]. As chromatin is an important interface for cellular processes such as transcription and DNA repair, it is not surprising that variants in this complex can lead to human disease [Ronan et al., 2013]. The BAF complex is also known as the SWI/SNF complex, but agreeing with the arguments of others [Ronan et al., 2013], we defer from using the SWI/SNF nomenclature to avoid bias and extrapolation.
In recent years somatic variants in several components of the BAF complex (such as ARID1A [MIM #603024], ARID1B, SMARCB1 [MIM #601607], and SMARCE1 [MIM #603111]) have been identified in many different types of tumors [Guan et al., 2011; Robinson et al., 2012; Samartzis et al., 2012; Stephens et al., 2012; Wang et al., 2011; Wiegand et al., 2010; Wilson and Roberts, 2011; Zang et al., 2012]. Truncating germline variants in SMARCA4 (MIM #603254) and SMARCB1 have been shown to cause rhabdoid tumor predisposition syndrome [Schneppenheim et al., 2010; Sevenet et al., 1999], and truncating germline variants in SMARCE1 were recently linked to familial multiple spinal meningiomas [Smith et al., 2013].
The first aim of our study was to define the genotype–phenotype relationship in CSS. Next generation sequencing techniques are widely used in research. Gradual introduction of targeted exome sequencing into diagnostic practice has been advocated [de Ligt et al., 2012; Rauch et al., 2012]. Thus, large numbers of known intellectual disability genes will be screened in individuals who present with learning difficulties, and variants of uncertain importance will be detected in individuals with phenotypes that do not allow immediate clinical recognition of a distinct entity. This necessitates extremely detailed “next-generation” genotype–phenotype studies as only these data will allow adequate counseling of families [Hennekam and Biesecker, 2012].
The second aim of our study was to estimate the mutational yield in a diagnostic setting. Patients with clinical features toward the severe end of the spectrum are typically selected for studies aiming at the identification of causative genes. In consequence, the mutation detection rate and genotype–phenotype correlations from such articles are often biased. Follow-up studies describing the results of genetic screening in a broader group of patients are essential for more definitive and accurate genotype–phenotype correlations.
Publication of additional pathogenic variants can provide insight into the mechanisms underlying the pathophysiological process and may facilitate the discrimination between nonpathogenic variants and pathogenic variants. The third aim of our study was to evaluate our ability to make this discrimination in the case of BAF complex genes. We extracted all variants previously identified from the Exome Variant Server to verify if these putatively non-pathogenic variants could rightly be classified as such or might have been classified as pathogenic variants if they would have been found in patients with intellectual disability.
Patients and Methods
Patients
Sixty three patients with a clinical diagnosis of CSS were enrolled. There are no widely accepted clinical diagnostic criteria for CSS. The vast majority of patients presented with a combination of intellectual disability, hypertrichosis, and fifth-finger abnormalities (either brachydactyly or small nails). Referring physicians were asked to complete a questionnaire concerning the clinical details. Informed consent was obtained to publish photographs. Patients 12 [Baban et al., 2008], 34 [Sousa et al., 2009; Van Houdt et al., 2012], and 101–103 [Santen et al., 2012a] were published previously.
Screening
All genes of the BAF subunits linked to CSS or NCBRS (ARID1A [NM_006015.4], ARID1B [NM_020732.3], SMARCA2 [NM_003070.3], SMARCA4 [NM_001128849.1], SMARCB1 [NM_003073.3], and SMARCE1 [NM_003079.4] were screened using conventional Sanger sequencing. All coding exons and intron/exon boundaries were included. Additionally, ARID1A and ARID1B were screened using custom-designed MLPA kits to detect exonic deletions and/or duplications, containing at least one probe per exon. Primer sequences, PCR conditions and MLPA probe sequences are available on request. PCR products were amplified using standard protocols. Sequencing reactions were performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Cheshire, UK) and separated on the ABI 3730 (Applied Biosystems). Results were analyzed using Sequence Pilot version 4.0.1 (JSI Medical Systems GmbH, Kippenheim, Germany).
Initially, all patients were screened for variants in ARID1B. If no pathogenic variant was identified, the other genes (ARID1A, SMARCB1, SMARCA2, SMARCA4, and SMARCE1) were screened in parallel. If sufficient DNA was present, screening of the other genes was also performed in those cases in which a pathogenic variant in ARID1B was identified. Since all pathogenic variants in SMARCA2 have thus far been identified in exons 15–25 [Van Houdt et al., 2012; Wolff et al., 2012], we analyzed only these exons for this study.
ARID1A- and ARID1B variants were considered pathogenic if they were truncating. For the other genes, missense variants were considered pathogenic when they included highly conserved nucleotides (as determined using phyloP), changed a highly conserved amino acid and were predicted to be possibly damaging by two out of three of the in silico prediction programs Polyphen2 [Adzhubei et al., 2010], SIFT [Ng and Henikoff, 2003], and MutationTaster [Schwarz et al., 2010]. De novo variants were all considered pathogenic.
Databases
All variants and clinical information were stored in the Leiden Open Variant Database (LOVD)-powered gene variant databases [Fokkema et al., 2011] for these genes (http://databases. lovd.nl/shared/).
Variants from Exome Variant Server
All documented variants in the BAF subunits were extracted from the Exome Variant Server (Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs. gs.washington.edu/EVS/) on February 23, 2013. The variants are summarized in Supp. Table S1.
Results
Sequencing of Patients
A pathogenic variant was found in 45 patients (71%) (Table 1, Figs. 1 and 2). The majority of pathogenic variants was identified in ARID1B (n = 28). For some cases (n = 8), DNA quality was not optimal, resulting in suboptimal amplification of the GC-rich exon 1 of ARID1A.
| Patient | Predicted protein | Amino acid | Functional | |||||
|---|---|---|---|---|---|---|---|---|
| ID | Gene | Exon | DNA variant | change | Inheritance | phyloP | conservation | predictions |
| 48 | ARID1A | 1 | c.1113del | p.Gln372Serfs*19 | De novo | |||
| 26 | ARID1A | 14 | c.3679G>T | p.Glu1227* | De novo | |||
| 33 | ARID1A | 20 | c.6493G>T | p.Glu2165* | Not in father | |||
| 25 | ARID1A | 20 | c.6532del | p.Asp2178Thrfs*22 | – | |||
| 24 | ARID1B | 1–20 | Whole gene deletion | p.0 | – | |||
| 20 | ARID1B | 1 | c.1222dup | p.Gln408Profs*127 | De novo | |||
| 55 | ARID1B | 1 | c.1235dup | p.Ser413Valfs*122 | De novo | |||
| 19 | ARID1B | 1 | c.1259dup | p.Asn420Lysfs*115 | De novo | |||
| 16 | ARID1B | 1 | c.1346del | p.Pro449Argfs*53 | De novo | |||
| 15 | ARID1B | 1 | c.1542+1G>A | r.spl? | De novo | |||
| 47 | ARID1B | 6–9 | Deletion of exon 6–9 | p.0 | De novo | |||
| 63 | ARID1B | 9 | c.2598del | p.Tyr867Thrfs*47 | – | |||
| 49 | ARID1B | 9 | c.2803dup | p.Met935Asnfs*7 | – | |||
| 65 | ARID1B | 9 | c.2877del | p.Ser959Argfs*9 | De novo | |||
| 57 | ARID1B | 10 | c.2998del | p.Ala1000Argfs*5 | De novo | |||
| 30 | ARID1B | 11 | c.3135+1G>C | r.spl? | Not in mother | |||
| 102 | ARID1B | 12 | c.3223C>T | p.Arg1075* | De novo | |||
| 10 | ARID1B | 15 | c.3846dup | p.Gly1283Trpfs*38 | De novo | |||
| 31 | ARID1B | 16 | c.4009C>T | p.Arg1337* | De novo | |||
| 12 | ARID1B | 17 | c.4098C>G | p.Tyr1366* | De novo | |||
| 51 | ARID1B | 18 | c.4448_4449ins14 | p.Pro1489Leufs*10 | – | |||
| 56 | ARID1B | 18 | c.4620C>A | p.Tyr1540* | De novo | |||
| 103 | ARID1B | 18 | c.4622_4631del | p.Gln1541Argfs*35 | De novo | |||
| 11 | ARID1B | 19 | c.4911_4915del | p.Trp1637Cysfs*6 | De novo | |||
| 101 | ARID1B | 20 | c.5329A>T | p.Lys1777* | De novo | |||
| 62 | ARID1B | 20 | c.5394_5397del | p.Phe1798Leufs*52 | De novo | |||
| 29 | ARID1B | 20 | c.5635delG | p.Asp1879Thrfs*95 | De novo | |||
| 34 | ARID1B | 20 | c.5968C>T | p.Arg1990* | Not in mother | |||
| 50 | ARID1B | 20 | c.5968C>T | p.Arg1990* | – | |||
| 73 | ARID1B | 20 | c.5968C>T | p.Arg1990* | De novo | |||
| 28 | ARID1B | 20 | c.6038G>A | p.Trp2013* | De novo | |||
| 39 | ARID1B | 20 | c.6233del | p.Pro2078Leufs*21 | De novo | |||
| 36 | ARID1B | 20 | c.6700_6701del* | p.Leu2234Glyfs*7a | –a | |||
| 13 | SMARCA2 | 18 | c.2554G>C | p.Glu852Gln | De novo | 6.3 | Tetraodon | SMP |
| 22 | SMARCA2 | 19 | c.2815C>T | p.His939Tyr | De novo | 6.1 | Tetraodon | SMP |
| 53 | SMARCA2 | 24 | c.3293G>A | p.Gly1098Asp | – | 6.2 | Tetraodon | SMP |
| 52 | SMARCA2 | 24 | c.3314G>A | p.Arg1105His | De novo | 6.2 | Tetraodon | SMP |
| 46 | SMARCA4 | 9 | c.1349C>A | p.Ala450Asp | De novo | 5.5 | Zebrafish | SMP |
| 70 | SMARCA4 | 23 | c.3127C>T | p.Arg1043Trp | De novo | 0.1b | Zebrafish | SMP |
| 42 | SMARCA4 | 25 | c.3380A>G | p.Asp1127Gly | Not in mother | 4.6 | Zebrafish | SMP |
| 2 | SMARCA4 | 27 | c.3608G>A | p.Arg1203His | – | 5.5 | Zebrafish | SMP |
| 43 | SMARCB1 | 8 | c.1089G>T | p.Lys363Asn | De novo | 1.3b | C. elegans | SMP |
| 5 | SMARCB1 | 8 | c.1091_1093del | p.Lys364del | De novo | – | C. elegans | – |
| 18 | SMARCB1 | 8 | c.1091_1093del | p.Lys364del | – | – | C. elegans | – |
| 37 | SMARCB1 | 8 | c.1091_1093del | p.Lys364del | De novo | – | C. elegans | – |
| 72 | SMARCE1 | 6 | c.314G>A | p.Arg105Gln | In affected mother | 6.3 | Tetraodon | M |
- –, In the inheritance columns indicates that parental DNA was not available for testing. Vertebrate phyloP score, species up to which the amino acid is conserved and functional predictions are given for missense variants. S, pathogenic according to SIFT; P, pathogenic according to Polyphen2; M, pathogenic according to MutationTaster.
- a The clinical significance of the c.6700_6701del variant is unclear, as it introduces a frameshift only 16 amino acids before the stop codon and parental DNA was not available for testing.
- b This base is the third base of a codon.
- Footnote for table: Used transcripts: ARID1A: NM_006015.4, ARID1B: NM_020732.3, SMARCA2: NM_003070.3, SMARCA4: NM_001128849.1, SMARCB1: NM_003073.3 and SMARCE1: NM_003079.4. The variants have been uploaded in the following LOVD databases: www.lovd.nl/ARID1A, www.lovd.nl/ARID1B, www.lovd.nl/SMARCA2, www.lovd.nl/SMARCA4, www.lovd.nl/SMARCB1, www.lovd.nl/SMARCE1.

ARID1A
In ARID1A four variants were identified which were truncating and therefore classified as pathogenic. The two nonsense substitutions were mosaic, and although it was less obvious for the frameshift variants, the variant peaks were consistently lower than the wild-type peaks, which suggest mosaicism (Supp. Fig. S1). In addition to DNA isolated from peripheral blood, we sequenced DNA isolated from cultured fibroblasts in one patient and found the variant in a much higher percentage (Supp. Fig. S1). We did not identify deletions in ARID1A by MLPA.
ARID1B
In ARID1B, 28 pathogenic truncating variants were identified (Table 1). In all cases where parental samples were available (n = 21), these variants arose de novo. We also identified a frameshift variant (c.6700_6701del;p.Leu2234Glyfs*7) in patient 36 near the 3′ end of the coding region (16 amino acids from the coding terminus). The status of this variant remains unclear because parental DNA was not available. We therefore decided not to include this patient in the phenotypic analysis. We also identified several missense variants and in-frame deletions and duplications in the repeat region in exon 1 (not reported), but in all cases, these were inherited from one of the healthy parents and therefore classified as nonpathogenic.
SMARCA2/SMARCA4
In the SMARCA2 and SMARCA4 genes, eight pathogenic variants were identified. All were missense variants, located at conserved positions in different functional domains. The variants arose de novo in all cases where parental DNA was available (n = 5). SMARCA2 and SMARCA4 encode the ATP-hydroxylase containing subunits of the BAF complex, and the missense variants are expected to influence the ATP-hydrolysis and therefore the function of this complex.
SMARCB1
All four pathogenic variants in SMARCB1 were identified in two adjacent amino acids. The variants arose de novo in all cases in which parental DNA was available (n = 3). One variant was found three times (c.1091_1093del; p.Lys364del) and the c.1089G>T;p.Lys363Asn variant was found in one patient.
SMARCE1
In SMARCE1, only a single possibly pathogenic missense variant was identified (c.314G>A;p.Arg105Gln). The variant is located in the high mobility group (HMG). The variant was inherited from the mother who has phenotypic similarities to her son. Grandparents or sibs of mother were not available for testing. Although the base and the amino acid are highly conserved (Table 1), the physicochemical distance between arginine and glutamine is small. Only MutationTaster predicts the change to be pathogenic.
Genotype–Phenotype Correlations
Comprehensive data on the clinical course and the results of the detailed phenotyping in the present series of patients are provided for each of the six groups of patients with pathogenic variants in one of the genes of the BAF chromatin remodeling complex (Table 2). All data used for this comparison may be found in Supp. Table S2. Photographs of all patients for which consent for publication was obtained are shown in Supp. Figure S2. The individual patient data are stored in the LOVD database (http://databases.lovd.nl/shared/). As pathogenic variants in SMARCA2 have thus far only been identified in patients with NCBRS, we have not taken the SMARCA2 patients into account when assessing the CSS phenotype. Although there were more females than males in our cohort (39 vs. 24, exact binomial test, P value 0.077), there was no gender effect in each of the patient groups (exact binomial test, P value > 0.5).
| Phenotype | ARID1A | ARID1B | SMARCA2 | SMARCA4 | SMARCB1 | SMARCE1 | No mutation |
|---|---|---|---|---|---|---|---|
| Height < −2.5 SD | 0% (0/4) | 11% (3/27) | 25% (1/4) | 25% (1/4) | 100% (4/4) | 0% (0/1) | 24% (4/17) |
| Weight < −2.5 SD | 25% (1/4) | 39% (11/28) | 50% (2/4) | 75% (3/4) | 50% (2/4) | 100% (1/1) | 41% (7/17) |
| OFC < −2.5 SD | 0% (0/3) | 0% (0/27) | 25% (1/4) | 25% (1/4) | 50% (2/4) | 0% (0/1) | 28% (5/18) |
| Clinical course | |||||||
| Feeding problems | 100% (4/4) | 64% (18/28) | 50% (2/4) | 100% (4/4) | 100% (4/4) | 100% (1/1) | 47% (8/17) |
| Hypotonia | 100% (4/4) | 85% (23/27) | 50% (2/4) | 75% (3/4) | 75% (3/4) | 0% (0/1) | 75% (12/16) |
| Seizures (including febrile, excluding abnormal EEG only) | 25% (1/4) | 20% (5/20) | 75% (3/4) | 0% (0/4) | 75% (3/4) | 100% (1/1) | 27% (4/15) |
| Vision problems | 100% (2/2) | 65% (17/26) | 66% (1/3) | 0% (0/4) | 75% (3/4) | 100% (1/1) | 75% (9/12) |
| Myopia (D) <-8, <-5, <-2, <0, none | 0-0-1-0-0 | 2-2-3-4-8 | 0-0-0-2-1 | 0-0-0-0-2 | 2-0-0-1-1 | 1-0-0-0-0 | 0-0-1-0-8 |
| Hearing problems | 33% (1/3) | 23% (5/22) | 0% (0/3) | 0% (0/4) | 33% (1/3) | 0% (0/1) | 7% (1/15) |
| Development | |||||||
| Delayed motor development (gross) | 100% (4/4) | 89% (25/28) | 100% (4/4) | 100% (4/4) | 100% (4/4) | 100% (1/1) | 73% (11/15) |
| Delayed motor development (fine) | 75% (3/4) | 95% (21/22) | 100% (2/2) | 33% (1/3) | 33% (1/3) | 100% (1/1) | 73% (8/11) |
| Cognition delay (mild–moderate–severe) | 1-0-2 | 3-16-9 | 1-2-1 | 0-2-2 | 0-0-4 | 0-1-0 | 3-6-6 |
| Speech delay (mild–moderate–severe) | 0-1-3 | 2-9-17 | 1-1-2 | 0-3-1 | 0-1-3 | 0-1-0 | 2-3-10 |
| Dysmorphic features | |||||||
| Sparse scalp hair | 0% (0/4) | 64% (18/28) | 100% (4/4) | 25% (1/4) | 75% (3/4) | 0% (0/1) | 72% (13/18) |
| Thick eyebrows | 50% (2/4) | 89% (24/27) | 100% (4/4) | 25% (1/4) | 100% (4/4) | 100% (1/1) | 67% (12/18) |
| Long eyelashes | 100% (2/2) | 82% (23/28) | 100% (4/4) | 50% (2/4) | 100% (4/4) | - | 67% (12/18) |
| Nonfunctioning or absent tear duct | 33% (1/3) | 20% (5/25) | 50% (1/2) | 33% (1/3) | 0% (0/2) | 0% (0/1) | 13% (1/8) |
| Flat nasal bridge | 100% (3/3) | 59% (16/27) | 50% (2/4) | 25% (1/4) | 100% (2/2) | 100% (1/1) | 67% (12/18) |
| Thick alae nasi | 67% (2/3) | 74% (20/27) | 100% (4/4) | 50% (2/4) | 100% (4/4) | 100% (1/1) | 72% (13/18) |
| Anteverted nose | 100% (4/4) | 52% (4/27) | 100% (4/4) | 50% (2/4) | 75% (3/4) | 0% (0/1) | 61% (11/18) |
| Broad philtrum | 67% (2/3) | 76% (19/25) | 75% (3/4) | 25% (1/4) | 67% (2/3) | 100% (1/1) | 88% (15/17) |
| Philtrum (long-short-normal) | 2-0-1 | 12-5-5 | 2-0-1 | 3-1-0 | 2-0-2 | - | 10-4-2 |
| Thick lower vermillion | 66% (2/3) | 92% (22/24) | 66% (2/3) | 75% (3/4) | 50% (2/4) | 100% (1/1) | 71% (12/17) |
| Large mouth | 33% (1/3) | 78% (21/27) | 100% (3/3) | 75% (3/4) | 75% (3/4) | 0% (0/1) | 94% (16/17) |
| Malformed ears | 100% (3/3) | 50% (14/28) | 75% (3/4) | 75% (3/4) | 100% (4/4) | 100% (1/1) | 50% (9/18) |
| Hypertrichosis | 75% (3/4) | 93% (26/28) | 100% (4/4) | 100% (4/4) | 100% (4/4) | 100% (1/1) | 83% (15/18) |
| Limb/trunk anomalies | |||||||
| Brachydactyly general | 66% (2/3) | 46% (11/24) | 33% (1/3) | 50% (2/4) | 25% (1/4) | 0% (0/1) | 53% (9/17) |
| Brachydactyly 5th finger | 66% (2/3) | 64% (16/25) | 50% (2/4) | 50% (2/4) | 75% (3/4) | 100% (1/1) | 69% (11/16) |
| Hypoplastic nails | 100% (4/4) | 68% (19/28) | 0% (0/2) | 100% (4/4) | 75% (3/4) | 0% (0/1) | 80% (12/15) |
| Hypoplasia or absence of 5th distal phalanx | 50% (2/4) | 23% (5/22) | 0% (0/3) | 66% (2/3) | 66% (2/3) | 0% (0/1) | 24% (4/17) |
| Prominent distal phalanges | 0% (0/2) | 38% (8/21) | 50% (2/4) | 0% (0/3) | 100% (3/3) | 100% (1/1) | 20% (3/15) |
| Prominent interphalangeal joints | 0% (0/2) | 35% (7/20) | 66% (2/3) | 33% (1/3) | 50% (2/4) | 100% (1/1) | 27% (4/15) |
| Joint laxity | 66% (2/3) | 75% (18/24) | 0% (0/4) | 100% (2/2) | 75% (3/4) | - | 60% (9/15) |
| Congenital anomalies | |||||||
| Laryngo- or tracheomalacia (# times reported) | 1 | 2 | - | 2 | - | - | 2 |
| Dextrocardia | 0% (0/4) | 0% (0/27) | 0% (0/4) | 0% (0/4) | 50% (2/4) | - | 0% (0/15) |
| Horseshoe kidney | 0% (0/3) | 0% (0/27) | 0% (0/4) | 0% (0/4) | 25% (1/4) | - | 0% (0/12) |
| Other | |||||||
| Delayed primary dentition | 100% (2/2) | 42% (8/19) | 50% (2/4) | 50% (2/4) | 50% (1/2) | 0% (0/1) | 18% (2/11) |
| Delayed permanent dentition | 100% (1/1) | 54% (7/13) | 33% (1/3) | 100% (1/1) | 50% (1/2) | 0% (0/1) | 0% (0/3) |
| Frequent infections | 66% (2/3) | 52% (12/23) | 33% (1/3) | 100% (3/3) | 50% (1/2) | 100% (1/1) | 64% (9/14) |
| Malignancies | 0% (0/3) | 0% (0/25) | 0% (0/4) | 0% (0/3) | 0% (0/4) | 0% (0/1) | 9% (1/11) |
| Radiological anomalies | |||||||
| Agenesis of the corpus callosum | 67% (2/3) | 36% (9/25) | 0% (0/3) | 100% (2/2) | 100% (3/3) | - | 36% (4/11) |
| Hypoplastic distal phalanx of fingers or toes | 50% (1/2) | 48% (11/23) | 0% (0/3) | 100% (1/1) | 100% (2/2) | 100% (1/1) | 58% (7/12) |
| Delayed bone age | 50% (1/2) | 17% (2/12) | 0% (0/3) | - | 100% (2/2) | - | 67% (4/6) |
- Some items were frequently spontaneously noted, these are indicated as # of times reported. Patient 36 was excluded from the analysis as the variant in ARID1B is of doubtful pathogenicity.
Discussion
Mutation Detection Rate
One aim of the study was to estimate the mutational yield of CSS screening in a diagnostic setting. In our cohort, we identified pathogenic variants in 71% (45/63) of patients, which is lower than previously reported (87%) [Tsurusaki et al., 2012]. By including all patients that were sent to us with a clinical diagnosis of CSS, without any further selection, we believe that our detection rate reflects the clinical reality more accurately, although some referral bias may still remain. The distribution of the pathogenic variants across the genes differs significantly from that published by Tsurusaki et al. ARID1B plays a more prominent role in our cohort (68% vs. 32%), and fewer pathogenic variants are identified in the other genes (Supp. Fig. S3). Variants in SMARCE1 seem to be a very rare cause of CSS, both in our study and in the literature.
The genes of all implicated subunits were sequenced in 53 patients. We did not identify more than a single pathogenic variant in any patient.
Discussion of Pathogenic Variants Per Gene
ARID1A/ARID1B
As the pathogenic variants in both ARID1A and ARID1B lead to haploinsufficiency, and size of the coding region is comparable (6,855 bp vs. 6,747 bp), we expected a priori to find a similar number of pathogenic variants in ARID1A and ARID1B. Clearly, this has now been proven wrong as we found four pathogenic variants in ARID1A compared with 28 in ARID1B. Although selection bias cannot be excluded, an appealing explanation is that truncating germline variants in ARID1A are embryonically lethal, especially since all pathogenic variants we identified in ARID1A appeared mosaic in lymphocytes (Supp. Fig. S1). We have meanwhile identified another patient with a pathogenic variant in ARID1A which appears mosaic (data not shown). No ARID1B-knockout mice have been described in the literature, but heterozygous truncating variants in ARID1A haven been shown to be embryonically lethal in mice [Gao et al., 2008]. Tsurusaki et al. (2012) showed the Sanger sequencing results of three variants in ARID1A, in which the mutant alleles of two variants had lower peaks than the reference allele. The c.31_56del; p.Ser11Alafs*91 variant at the start of exon 1 has equal peak heights, which may be explained by (a) the variant is nonmosaic in blood, but mosaic in other tissue, (b) the alternative transcript (transcript ID uc001bmw.1, Ensembl transcript ID ENST0000037415, which lacks exon (1) is more important than the canonical transcript, or (c) there is an alternative translation start site, and the encoded protein has sufficient functionality to be nonlethal. As the c.31_56del variant was not tested in parental DNA, we cannot exclude that the variant is in fact benign.
We identified two deletions in ARID1B and none in ARID1A. This low number of larger deletions may be because most referring clinicians had sought copy number variation using array techniques, although these might not detect small deletions. The limited sensitivity of MLPA in detecting mosaicism may also play a role for ARID1A.
Although in the present study no RNA analyses were performed, Tsurusaki et al. have shown that the c.5632del; p.Asp1878Metfs*96 variant in the last exon of ARID1B seems to escape nonsense mediated decay. This may suggest that pathogenic variants in exon 20 result in a truncated protein and may point toward the presence of protein domains in the 3′ coding end essential for ARID1B function, as for example the BC-box identified in c.6244_6273 [Li et al., 2010]. However, without protein analysis one cannot exclude that the truncated protein or RNA is unstable leading to pure haploinsufficiency.
We found no evidence that missense variants in ARID1B might lead to haploinsufficiency in a recessive manner in our current study or in literature. Only variants leading to haploinsufficiency were considered as pathogenic when we reclassified the EVS data set. No such variants were present in ARID1A. In ARID1B, several frameshift variants in the EVS data might be considered pathogenic (Supp. Tables S1 and S3). The majority of these variants are located in the first exon. As the coverage reported in the EVS database is low in exon 1 (due to the high GC content of this exon) these frameshift variants may be artifacts. Similar to others [Hoyer et al., 2012], we have identified several in-frame duplications and deletions in our patients in exon 1, all of which were shown to be inherited from healthy parents if parental DNA was available. Such variants might contribute to artificial frameshift variants in the context of next generation sequencing variant calling.
An alternative explanation might have been that these variants do not lead to haploinsufficiency because transcription is rescued by an alternative starting codon. As we identified several de novo variants in exon 1, we rejected this explanation.
The c.6008del variant in the EVS is located in exon 20. Both we and Hoyer et al. (2012) (c.6463_6473del) have identified several de novo variants distal of this position, and as the c.6008del variant was reported only once in the EVS, it may be a false-positive. Alternatively, the individual with this variant might represent the mildest end of the phenotypic spectrum of ARID1B patients. As one of our patients has low-normal cognitive ability this possibility cannot be ruled out.
SMARCA2/SMARCA4
SMARCA2 and SMARCA4 are mutually exclusive in the BAF complex [Hargreaves and Crabtree, 2011]. They both have an important function in the hydrolysis of ATP to generate energy to remodel the chromatin. All variants linked to intellectual disability are missense variants in highly conserved functional domains of the protein. Haploinsufficiency of SMARCA4 causes an increased susceptibility to rhabdoid tumors (Rhabdoid Tumor Predisposition Syndrome) [Schneppenheim et al., 2010]. Haploinsufficiency of SMARCA2 has not been linked to human disease. We have hypothesized [Santen et al., 2012b] that the reason for such differing phenotypes between missense and nonsense variants is that missense variants may lead to properly constituted but nonfunctional BAF complex interfering with wild-type BAF complex, leading to a dominant negative effect by competitive inhibition.
The EVS contained 36 and 44 missense variants in SMARCA2 and SMARCA4, respectively. Three missense variants in SMARCA2 are located in the exons 15–25 (Supp. Table S3), which contain the ATP-hydroxylase domains. Each of these variants was found in a single individual, and all of them are highly conserved and predicted to be pathogenic by at least two protein prediction programs. In SMARCA4, 9/44 missense variants were located in annotated protein domains, and seven of these are found in only a single patient (Supp. Table S3). Therefore, these variants may represent false-positive findings or provide evidence that it is difficult to separate nonpathogenic from pathogenic missense variants in SMARCA2 and SMARCA4.
Canonical splice-site variants (one in SMARCA2 and five in SMARCA4) were also found in the EVS (Supp. Table S1). The c.1347+1G>A variant in SMARCA2 is predicted to cause an in-frame skip of exon 7. Exon 7 does not contain an annotated domain and we concluded that this variant is not pathogenic. The same holds for c.1118+1_1118+2insC in SMARCA4, which might also cause a skip of exon 7. The c.2506–2dup variant is predicted to have only a minor effect on the splice acceptor site of exon 19, and is therefore probably nonpathogenic. The other splice-site variants are all predicted to lead to haploinsufficiency, and thus to rhabdoid tumor predisposition syndrome rather than intellectual disability.
SMARCB1
Although SMARCB1 is an established core component of the BAF complex [Wilson and Roberts, 2011], its precise function is not clear. It does not appear to have a DNA-binding domain, and probably acts as some sort of regulator or stabilizer of the complex. Interestingly, pathogenic variants in very few positions of SMARCB1 have been identified in CSS [Tsurusaki et al., 2012], although one de novo variant was described in exon 2 in a patient with a clinical diagnosis of Kleefstra syndrome [Kleefstra et al., 2012]. Amino acids 363, 364, and 377 seem to be vital in the proper function of the BAF complex. As amino acids 352–378 are highly conserved new pathogenic variants may be expected there. Uniprot does not annotate a functional domain for these amino acids, indicating that variants in the other exons might yet be pathogenic. Like SMARCA4, haploinsufficiency of SMARCB1 causes rhabdoid tumor predisposition syndrome [Sevenet et al., 1999]. In addition, haploinsufficiency of SMARCB1 causes schwannomatosis in some families [Swensen et al., 2009].
All SMARCB1 variants in the EVS are classified as probably nonpathogenic in the context of CSS as they are not close to the thus far implicated amino acids. It is worth noting that only four missense variants are reported in the EVS in the other exons of SMARCB1, each of them present in a single patient.
SMARCE1
The c.314G>A variant we identified in the HMG domain of SMARCE1 in a patient and his mother with a similar phenotype remains of doubtful pathogenicity. The earlier reported SMARCE1 variant [Tsurusaki et al., 2012] is more conserved and the difference between the amino acids is larger (tyrosine to cysteine). All prediction programs predict this variant to be pathogenic.
The difficulty in classifying SMARCE1 variants is further illustrated by examination of the EVS test data set. Of the 15 missense variants in SMARCE1, 11 were observed in only a single patient in the EVS, and one was observed in ≥3 patients. This may signify an enrichment of false-positive variants. Only one of these variants was located in the HMG domain (c.330A>C), and is in many respects similar to the possibly pathogenic variant we identified. It has the same predictions by the protein prediction programs, but is slightly less conserved (phyloP 2.5). This may be due to the base being the third base in the codon. However, as very few pathogenic variants in SMARCE1 are known, it remains possible that variants outside the HMG domain are pathogenic.
Genotype–Phenotype Correlation
General Conclusions About the CSS Phenotype
We have identified manifestations that are present in most individuals with CSS and others that are specific to pathogenic variants in one particular gene (Supp. Table S2). The most consistent manifestations, present in at least half of all patients with a pathogenic variant in any of the genes (excluding SMARCA2, as discussed below), are intellectual disability (98%), hypotonia (83%), feeding problems (76%), hypertrichosis (93%), sparse scalp hair (61%), thick eyebrows (92%), long eyelashes (82%), visual problems (62%), thick alae nasi (74%), large mouth (72%), thick lower vermillion (83%), malformed ears (63%), lax joints (76%), short fifth finger (65%), and one or more underdeveloped nails (76%). We consider these the core manifestations of CSS. Some of the findings, especially sparse hair, are more frequently seen in younger patients, with many having sparse scalp hair in infancy which resolves during childhood. Microcephaly has been reported as a sign of CSS [Schrier et al., 2012], but was uncommon in the present series. Underdevelopment or absence of the corpus callosum has been previously linked to CCS [Halgren et al., 2012; Santen et al., 2012a] but was observed here in only a minority of the patients.
Few organ malformations were found in the present series of patients. The finding of dextrocardia in two of the SMARCB1 patients and a horseshoe kidney in another SMARCB1 patient are the exceptions. This contrasts with earlier reports [Schrier et al., 2012].
Visual problems in the present series consisted mainly of severe myopia, up to −18D. Hearing problems were much less common and mainly explainable by recurrent upper airway infections.
Somatic and germline variants in BAF subunits have been linked to cancer [Wilson and Roberts, 2011]. There are no reports of individuals with CSS and a malignancy in the literature. In the present series, none of the molecularly confirmed patients had a malignancy; one of the patients without a detectable pathogenic variant had a clear cell carcinoma of the cervix uteri at age 20 years. Overall this would seem to indicate a low risk for developing a malignancy, although the short median follow-up in the present series (10 years) prevents a definitive conclusion to be drawn.
Manifestations per gene involved
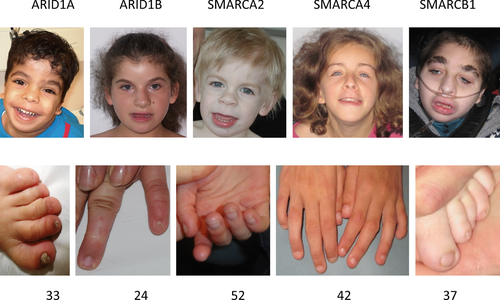
ARID1B patients (n = 28) usually do not have a significantly impaired prenatal or postnatal growth, and have normal brain growth, inferred by normal skull circumferences, moderate feeding problems in two-thirds, seizures in one-thirds, and also an underdeveloped corpus callosum in one-third of cases. In six patients, a scoliosis developed. We found hypertelorism and anteverted nares more commonly than in the other groups. Marked fetal pads were common, but thickened distal phalanges did not occur. Several patients did have long fingers, and a few patients showed an ulnar deviation of the fifth finger. If nails were small this was almost always confined to the fifth rays, and nails on other fingers or toes were of normal size, although these could be dystrophic.
ARID1A patients (n = 4) tended to have moderately to severely impaired development and feeding problems but height and head circumference were usually normal. Seizures were uncommon. They did not show significantly sparse hair but did have hypertelorism, anteverted nares, and a thin upper vermillion. Nails of all fingers and toes were often small but those on the fifth rays were very small. Unusual findings in just one patient were Hirschsprung disease, narrow choanae, and thoracic vertebral anomalies.
SMARCA4 patients (n = 4) had less marked feeding problems and their intellectual disability was considered moderate to severe. Head circumference was around the −2SD as was growth in height. They showed no seizures and had normal vision. Sparse hair was present but only mildly so, and their nares were not anteverted. The distal phalanges of the toes and fingers were not thickened, nails of the fifth rays were small but less compared with ARID1A patients and the other nails were only mildly decreased in size.
Individuals with pathogenic variants in SMARCB1 (n = 4) all showed reduced growth in height and half of them had reduced weight, and often had a microcephaly of around −3SD. Developmental delay was severe, feeding problems were more marked than in ARID1B-patients and had a more prolonged course. Multifocal seizures were common and severe, and have been fatal in one patient. Unusual manifestations were gynecomastia and alacrima, each in one patient. The phenotype is characterized by often sparse scalp hair, very thick eyebrows, and common changes in the nose, philtrum, and mouth. Fetal pads were less marked in this group, and fingers could be long. Decrease in nail size was confined to the fifth rays.
The emerging phenotype–genotype correlation suggests that SMARCB1 patients have the most severe delay, growth retardation, and most distinctive phenotype, whereas SMARCA4 and ARID1B patients are less severely affected. The variability in phenotype seems most marked in ARID1A and ARID1B patients, which may be explained at least in part in ARID1A by their mosaicism. The distal limb anomalies of especially the fifth rays are most pronounced in individuals with a pathogenic variant in ARID1A and least expressed in SMARCB1 patients. Numbers of individuals with pathogenic variants in each of the genes studied here are small and although indicative, no definitive conclusions on the phenotype can be drawn.
Patients without a detectable pathogenic variant presented similarly to those with a pathogenic variant, which is to be expected as these individuals were all clinically diagnosed as having CSS. Differences may be the less frequent myopia and less common hypertrichosis and joint laxity in the group without a pathogenic variant and more frequent general brachydactyly in the latter. The presence of core manifestations associated with CSS was similar in both groups, although feeding problems were usually less marked in those without pathogenic variants. We hypothesize that either mosaicism for the genes studied in the present series, as recently shown for Cornelia de Lange syndrome [Huisman et al., 2013], or variations in other genes in the BAF complex might be identified to be the cause of the phenotype in this group of patients.
In the present study, we enrolled only patients with a CSS clinical phenotype. Pathogenic variants in ARID1B may also, however, lead to intellectual disability without a recognizable phenotype [Halgren et al., 2012; Hoyer et al., 2012; Santen et al., 2012a] and such individuals would not have been ascertained in our study. It remains to be seen how many ARID1B patients actually have CSS.
In the current series of patients referred with a clinical diagnosis of CSS, we have identified four pathogenic SMARCA2 variants. Pathogenic missense variants in the ATP-helicase domains in SMARCA2 (exons 15–25) have been, until now, identified only in NCBRS patients [Van Houdt et al., 2012; Wolff et al., 2012]. Tsurusaki et al. reported a pathogenic SMARCA2 variant in their CSS cohort, but retrospectively reclassified this patient as having NCBRS (N. Matsumoto, personal communication).
The phenotype in NCBRS is considered to be specific and separate from the phenotype of CSS [Sousa et al., 2009]. Main points of difference are the markedly sparse hair that becomes sparser with time, the progressive coarseness of the face, thinning of subcutaneous tissues in the face, markedly broadened and thickened distal phalanges and normal nails. Unfortunately, we had too limited photographic material to reevaluate fully the phenotype of our four patients, but available pictures were compatible with NCBRS. The literature data and the larger series of SMARCA2 patients known to one of us (R.C. Hennekam, unpublished) emphasize the clear phenotypic distinction between SMARCA2 (NCBRS) patients and patients with pathogenic variants in the other five BAF subunits who have CSS. Consequently, it is more likely that the four patients have in fact NCBRS, but our findings underline that distinguishing between the two conditions may be difficult. We recommend screening of SMARCA2 if the clinical diagnosis of CSS is uncertain and the manifestations show overlap with NCBRS.
CSS as Clinical Entity
There has been debate amongst clinicians as to whether CSS represents a distinct clinical entity. The identification of pathogenic variants in ARID1B in individuals with nonsyndromic intellectual disability has led to further confusion [Halgren et al., 2012; Hoyer et al., 2012; Santen et al., 2012a]. Indeed, OMIM states (accessed March 16, 2013) that “patients diagnosed with Coffin–Siris syndrome have such broad clinical variability that Coffin–Siris syndrome no longer appears to be a distinct entity.” The results of the present study argue against this statement. We demonstrate a similar pattern of clinical features in a large group of individuals recognized clinically as having CSS, with subsequent more detailed phenotyping showing limited distinctions between the various subgroups; all subgroups are classified by genes that act in the same complex. In our opinion, this allows the definition of a genuine entity [Hennekam, 2007]. A similar situation pertains in other entities caused by defects in different components of the same complex such as Cornelia de Lange syndrome and Bardet–Biedl syndrome. Although pathogenic ARID1B variants have been identified in 0.9% of a large cohort of individuals with nonsyndromic intellectual disability [Hoyer et al., 2012], this still does not argue against CSS as a genuine entity, as the percentage of mutations is so much higher in CSS patients (44%), and similar situations are now occurring with other genes such as TCF4 and RSK2 [Rauch et al., 2012; Maystadt et al., 2013].
Clinical Recommendations
- Special attention should be given to seizure-like episodes. If there is any doubt an EEG should be performed.
- Vision and hearing should be screened at diagnosis, and additionally if parents later indicate the child does not see or hear well.
- The weight of patients should be monitored, and if it falls away from growth percentiles referral to a dietician should be organized. Gastrostomy should be considered in those with severe nutritional problems.
- There does not seem to be an increased risk of malignancies. Although some malignancies have been reported in literature, a screening program is at present not warranted.
Conclusions
We have presented a cohort of 63 patients with a clinical diagnosis of CSS with a pathogenic variant in 45 (71%) of the patients. We reclassified four SMARCA2 patients as NCBRS. The remaining yield is 69% (41/59). All our clinical and molecular data were stored in an LOVD3 database to facilitate usage and updating of the data. As a novel approach, we have assessed variants in the BAF complex genes present in the Exome Variant Server.
Using the EVS variants as a test data set we show that variants can be reclassified reliably for most genes. However, for SMARCA2, SMARCA4, and SMARCE1, it is difficult to discriminate between nonpathogenic and pathogenic variants without access to parental DNA, and we recommend analysis of parental samples. Functional studies will need to be performed to improve our ability to classify variants in these genes.
We present a genotype–phenotype correlation confirming CSS as a distinct entity and provide several recommendations for the clinical care of CSS patients that follow from our phenotypic data.
Acknowledgments
We would like to thank the patients and their parents for their cooperation, and Ivo Fokkema for his help in storing the clinical information in LOVD.
Disclosure statement: The authors declare no conflict of interest.
Appendix
Mariam Almureikhi (Section of Clinical and Metabolic Genetics, Department of Pediatrics, Hamad Medical Corporation, Doha, Qatar); Anwar Baban (Medical and Surgical Department of Pediatric Cardiology, Bambino Gesù Pediatric Hospital, Rome, Italy); Mafalda Barbosa (Centro de Genética Médica Jacinto Magalhães – Centro Hospitalar do Porto, Porto, Portugal; Instituto Gulbenkian de Ciência, Oeiras, Portugal); Tawfeg Ben-Omran (Clinical and Metabolic Genetics Division, Department of Pediatrics, Hamad Medical Corporation, 3050, Doha, Qatar); Katherine Berry (Department of Medical Genetics, Shodair Hospital, Helena, MT, USA); Stefania Bigoni (Medical Genetic Unit, Ferrara University Hospital, Ferrara, Italy); Odile Boute (CLAD Nord de France, Service de Genetique Clinique Guy Fontaine, CHRU, Lille, France); Louise Brueton (Birmingham Women's National Health Service (NHS) Foundation Trust, Birmingham, UK); Ineke van der Burgt (Department of Human Genetics, Radboud University Medical Centre, Nijmegen, The Netherlands); Natalie Canham (North West Thames Regional Genetics Service, Northwick Park Hospital, Harrow, UK); Kate E. Chandler (Department of Genetic Medicine, St Mary's Hospital, Manchester Academic Health Sciences Centre (MAHSC), University of Manchester, Manchester, UK); Krystyna Chrzanowska (Department of Medical Genetics, The Children's Memorial Health Institute, Warsaw, Poland); Amanda L. Collins (Wessex Clinical Genetics Service Princess Anne Hospital Coxford Road Southampton, UK); Teresa de Toni (Department of Pediatrics, University of Genova, Genova, Italy); John Dean (Medical Genetics, Ashgrove House, Foresterhill, Aberdeen, United Kingdom); Nicolette S. den Hollander (Center for Human and Clinical Genetics, Leiden University Medical Centre, Leiden, The Netherlands); Leigh Anne Flore (Division of Genetic & Metabolic Disorders, Department of Pediatrics, Children's Hospital of Michigan, Detroit, MI, USA); Alan Fryer (Department of Clinical Genetics, Liverpool Women's NHS Foundation Trust, Liverpool, UK); Alice Gardham (North West Thames Regional Genetics Service, Northwick Park Hospital, Harrow, UK); John M. Graham, Jr. (Medical Genetics Institute, Cedats-Sinai Medical Center, Los Angeles CA); Victoria Harrison (Department of Clinical Genetics, Churchill Hospital, Oxford, UK); Denise Horn (Institute for Medical and Human Genetics, Charité Universitätsmedizin Berlin, Berlin, Germany); Marjolijn C. Jongmans (Department of Human Genetics, Radboud University Medical Centre, Nijmegen, The Netherlands); Dragana Josifova (MD, FRCP, MRCPCH, Consultant in Clinical Genetics, Clinical Genetics Department, Guy's Hospital, London, UK); Sarina G. Kant (Center for Human and Clinical Genetics, Leiden University Medical Centre, Leiden, The Netherlands); Seema Kapoor (Pediatrics Genetic & Research Laboratory, Department of Pediatrics, Maulana Azad Medical College, New Delhi, India); Helen Kingston (Manchester Centre for Genomic Medicine, St Mary's Hospital, CMFT, Manchester, UK); Usha Kini (Department of Clinical Genetics, Oxford Radcliffe University Hospitals NHS Trust, Oxford, UK); Tjitske Kleefstra (Department of Human Genetics, Radboud University Medical Centre, Nijmegen, The Netherlands); Małgorzata Krajewska-Walasek (Department of Medical Genetics, The Children's Memorial Health Institute, Warsaw, Poland); Nancy Kramer (Cedars-Sinai Medical Center, Los Angeles, CA, USA); Saskia M. Maas (Departments of Pediatrics and Clinical Genetics, Academic Medical Center, Amsterdam, The Netherlands); Patricia Maciel (Life and Health Sciences Research Institute (ICVS), School of Health Sciences, University of Minho, Braga, Portugal and ICVS/3B's – PT Government Associate Laboratory, Braga/Guimarães, Portugal); Grazia M.S. Mancini (Department of Clinical Genetics, Erasmus University Medical Center, Rotterdam, The Netherlands); Isabelle Maystadt (Centre de Génétique Humaine, Institut de Pathologie et de Génétique, Avenue G. Lemaître 25, Charleroi, Belgium); Shane McKee (Northern Ireland Regional Genetics Service, Belfast City Hospital, Belfast, UK); Jeff M. Milunsky (Center for Human Genetics, Inc., Cambridge, MA, USA); Sheela Nampoothiri (Department of Pediatric Genetics, Amrita Institute of Medical Sciences and Research Center, Kerala, India); Ruth Newbury-Ecob (Department of Clinical Genetics, St Michael's Hospital, Bristol, UK); Sarah M. Nikkel (Department of Genetics, Children's hospital of Eastern Ontario, Ottawa, Ontario, Canada); Michael J. Parker (Sheffield Clinical Genetics Services, Sheffield Children's NHS Foundation Trust, UK); Luis A. Pérez-Jurado (Unitat de Genètica, Universitat Pompeu Fabra, Hospital del Mar Research Institute (IMIM), and Centro de Investigación en Red de Enfermedades Raras (CIBERER), Barcelona, Spain); Stephen P. Robertson (Department of Paediatrics, Dunedin School of Medicine, University of Otago, North Dunedin, New Zealand); Caroline Rooryck (Univ. Bordeaux, Maladies Rares : Génétique et Métabolisme (MRGM), EA 4576, F-33000, Bordeaux, France); Debbie Shears (Department of Clinical Genetics, Oxford Radcliffe University Hospitals NHS Trust, Oxford, UK); Margherita Silengo (Department of Pediatrics, University of Turin, Turin, Italy); Ankur Singh (Pediatrics Genetic & Research Laboratory, Department of Pediatrics, Maulana Azad Medical College, New Delhi, India); Robert Smigiel (Genetics Department Wroclaw Medical University, Wroclaw, Poland); Gabriela Soares (Centro de Genética Médica Jacinto Magalhães – Centro Hospitalar do Porto, Porto, Portugal); Miranda Splitt (Northern Genetics Service, Newcastle upon Tyne, UK); Helen Stewart (Department of Clinical Genetics, Oxford Radcliffe University Hospitals NHS Trust, Oxford, UK); Elizabeth Sweeney (Department of Clinical Genetics, Liverpool Women's NHS Foundation Trust, Liverpool, UK); May Tassabehji (Department of Genetic Medicine, University of Manchester, Oxford Road, Manchester, UK); I (Karen) Temple (Human Genetics and Genomic Medicine, Faculty of Medicine, University of Southampton, Southampton, UK); Beyhan Tuysuz (Department of Pediatric Genetics, Cerrahpaşa Medical School, Istanbul University, Istanbul, Turkey); Albertien M. van Eerde (Department of Medical Genetics, University Medical Center Utrecht, Utrecht, The Netherlands); Catherine Vincent-Delorme (Clad Nord de France-Service de Génétique Clinique Guy Fontaine CHRU Lille- Cs de Génétique CH Arras); Louise C. Wilson (Clinical Genetics, Great Ormond Street Hospital, London, UK); Gozde Yesil (Bezmialem University of Medicine, Department of Medical Genetics).




