Androgen Receptor Enhances Hepatic Telomerase Reverse Transcriptase Gene Transcription After Hepatitis B Virus Integration or Point Mutation in Promoter Region
Abstract
The gender disparity of hepatocellular carcinoma (HCC) is most striking in hepatitis B virus (HBV)-related cases. The majority of such HCC cases contain integrated HBV, and some hotspot integrations, such as those in the telomerase reverse transcriptase gene (TERT) promoter, activate gene expression to drive carcinogenesis. As the HBV genome contains both androgen-responsive and estrogen-responsive motifs, we hypothesized that the integrated HBV DNA renders a similar regulation for downstream gene expression and thus contributes to male susceptibility to HCC. To test this hypothesis, the HBV integration sites and the common mutations in the TERT promoter and tumor protein P53 (TP53) coding region were analyzed in 101 HBV-related HCC cases using a capture-next-generation sequencing platform. The results showed that both HBV integration and –124G>A mutation in the TERT promoter region, occurring in a mutually exclusive manner, were more frequent in male than in female patients with HCC (integration: 22/58 male patients with HCC, 6/36 female patients with HCC, P = 0.0285; –124G>A: 17/62 male patients with HCC, 3/39 female patients with HCC, P = 0.0201; in combination, 39/62 male patients with HCC, 9/39 female patients with HCC, P < 0.0001). The effects of sex hormone pathways on the expression of TERT with both genetic changes were investigated using a reporter assay. HBV integration in the TERT promoter rendered the TERT transcription responsive to sex hormones, with enhancement by androgen receptor (AR) but suppression by estrogen receptor, both of which were dependent on hepatocyte nuclear factor 4 alpha. Besides, AR also increased TERT expression by targeting TERT promoter mutations in a GA binding protein transcription factor subunit alpha–dependent manner. Conclusion: TERT elevation by AR through integrated HBV and point mutation at the TERT promoter region was identified as a mechanism for the male dominance of HBV-related HCCs; telomerase and AR thus may be targets for intervention of HCC.
Abbreviations
-
- AR
-
- androgen receptor
-
- ARE
-
- androgen response element
-
- BBAP
-
- BLAST-based assembly pipeline
-
- BLAST
-
- Basic Local Alignment Search Tool
-
- CCNE1
-
- cyclin E1 gene
-
- CDS
-
- coding sequence
-
- EnhI
-
- enhancer I
-
- ERE
-
- estrogen response element
-
- ERα
-
- estrogen receptor alpha
-
- GABPA
-
- GA binding protein transcription factor subunit alpha
-
- gDNA
-
- genomic DNA
-
- HBV
-
- hepatitis B virus
-
- HCC
-
- hepatocellular carcinoma
-
- HNF4α
-
- hepatocyte nuclear factor 4 alpha
-
- Luc
-
- luciferase
-
- MLL4
-
- mixed-lineage leukemia 4 gene
-
- NGS
-
- next-generation sequencing
-
- NTUH
-
- National Taiwan University Hospital
-
- si
-
- small interfering
-
- TERT
-
- telomerase reverse transcriptase gene
-
- TLCN
-
- Taiwan Liver Cancer Network
-
- TP53
-
- tumor protein P53
-
- WT
-
- wild type
Hepatocellular carcinoma (HCC), the third leading cause of cancer death,1 is a devastating complication of chronic liver diseases that is mainly caused by infection with hepatitis B virus (HBV) or hepatitis C virus, or by nonalcoholic fatty liver disease.2-4 Overall, men are more likely to develop HCC than women, with a tumor incidence ranging from 3.7 to 31.9/100,000 person-years in men and from 1.4 to 10.2/100,000 person-years in women.1 This gender difference is more striking in HBV-related HCC, with a male-to-female ratio of ~6:1.5 Notably, in HBV carriers, higher androgen pathway activity correlates with a higher incidence of HCC in men,6 while deprivation of estrogen due to menopause or oophorectomy increases HCC incidence in women.7 These findings suggest that sex steroids could be involved in HBV-induced carcinogenesis, functioning in an opposite manner by gender.
Our previous studies have shown that HBV is, in fact, a sex dimorphic virus. The androgen pathway promotes HBV transcription through the androgen response element (ARE) in viral enhancer I (EnhI).8 In contrast, the estrogen pathway inhibits HBV transcription by passively quenching essential hepatocyte nuclear factor 4 alpha (HNF4α) from binding to the cognate site within EnhI.9 The results provide a mechanism for the inverse effects of sex hormones in HBV transcription and replication, leading to increased male susceptibility to hepatocarcinogenesis.
In addition to being present in an episomal form, HBV exists as an integrated form in the chromosomes of some virus-infected hepatocytes, and approximately 90% of HBV-related HCCs contain integrated HBV DNA in their chromosomes.10, 11 Recent next-generation sequencing analysis of HBV-related HCC genomes not only confirmed frequent HBV integrations but also revealed a few integration hotspots close to the telomerase reverse transcriptase (TERT), mixed-lineage leukemia 4 (MLL4/KMT2B), and cyclin E1 (CCNE1) oncogenes.10, 12 The presence of integration hotspots in the chromosomes of HBV-related HCCs supports their roles in “insertional mutagenesis” to drive carcinogenesis.13, 14 Thus, HBV integration renders the adjacent cellular genes under the transcriptional control of HBV EnhI and therefore increases their expression levels. In cases in which the flanking cellular genes are oncogenic, this mechanism provides the hepatocyte with a great growth advantage for clonal expansion and eventually selects for HCC.10
According to this insertional mutagenesis mechanism elicited by the integrated HBV genome, we propose that the sex hormones may also affect the integrated HBV DNA, similar to the episomal one, to regulate the transcription of downstream genes and contribute to HCC development. This hypothesis was tested by comparing the HBV integration sites in male and female patients with HCC by extensive capture-NGS analysis. The results could help clarify whether certain cellular oncogenes activated by HBV insertion are enriched in male patients with HCC. Additionally, the roles of the androgen and estrogen pathways in regulating these gene expression levels were investigated accordingly. The results could provide a mechanism for the male dominance of HBV-related HCCs and even suggest targets for future HCC therapies.
Patients and Methods
Study Subjects
Tumor and nontumor paired liver tissues from 70 HBV-related HCC patients (46 men and 24 women) were collected at National Taiwan University Hospital (NTUH). The liver tissues were stored in liquid nitrogen after being resected for subsequent genomic DNA (gDNA) and RNA extraction. Another 31 pairs of tumor–nontumor gDNA and RNA from 16 male and 15 female patients with HBV-related HCC were obtained from the Taiwan Liver Cancer Network (TLCN). The institutional review boards of NTUH and TLCN approved the use of these archived samples. The clinical characteristics of these 101 patients are summarized in Supporting Table S1.
Capture-NGS Analysis
gDNA was extracted from the liver tissues using a standard phenol-chloroform method, and 1 µg of the gDNA was used for bar-coded library construction with the TruSeq DNA Sample Preparation kit (Illumina, San Diego, CA). The libraries were processed for hybridization with the customized biotinylated DNA probes (IDT, Coralville, IA) together with human Cot-1 DNA (Invitrogen, Carlsbad, CA) at 65°C for 4 hours, following the manufacturer’s instructions (SeqCap Hybridization and Wash Kit; Roche, Mannheim, Germany). After hybridization, the captured DNA was purified using Dynabeads M-270 Streptavidin (Invitrogen), and unbound DNA was removed with washing buffer. The target DNA fragments were amplified by PCR with the adapter primer set (Illumina) and KAPA HiFi HotStart ReadyMix (KAPA, Wilmington, MA). The PCR products were then purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA) and sequenced by HiSeq 2000 (Illumina).
Capture Probe Set
The capture probe set consisted of 200 biotinylated DNA oligonucleotide probes (IDT) covering the entire HBV genome and selected endogenous genetic regions (TERT promoter and coding region of tumor protein P53 [TP53]). The HBV probes were designed using HBV consensus sequences of genotypes B and C assembled from HBV whole-genome sequences that were sampled and sequenced from hepatitis B patients at NTUH. The National Center for Biotechnology Information reference sequences of selected endogenous gene regions, including –279 to +303 relative to ATG of the TERT gene (NC_000005.9) and exons 2-11 (total coding sequence) of TP53 (NM_000546.5), were used to design the endogenous gene probes.
Bioinformatics Analysis
Paired NGS data sets were treated as single reads and merged into single data sets. Quality control of the NGS data sets includes removal of low-quality reads, probe hybridization analysis, and exclusion of low-redundancy reads. Low-quality reads were defined as reads with a number of low-quality nucleotide sites (quality score <5) equal to or greater than half the read length and were excluded from further analyses. NGS data sets of high-quality/high-redundancy reads were compared against both HBV and human reference sequences by the Basic Local Alignment Search Tool (BLAST). The sequence reads containing the junction regions of HBV integration into the human genome were identified if one portion of the reads was of HBV origin and the remaining nonoverlapping portion was of human origin. This analysis was accomplished by initially comparing the sequence reads against the HBV reference sequences and then identifying partial HBV reads with a maximum HBV length of 100 bp and an e-value threshold of 1e-10. Partial HBV reads were then compared with the human hg19 reference sequences, and junction-containing reads were identified with an e-value threshold of 1e-10. Junction reads were clustered according to the pairwise BLAST results using BLAST-based assembly pipeline (BBAP)15 to identify unique junction types and to quantify the number of junction reads for each unique junction type. Junctions supported by two or more junction types and 15 or more junction reads were identified as viral/host (vh)–chimera DNA. High-quality/high-redundancy reads that were not identified as partial or full HBV reads were then assembled onto endogenous gene reference sequences consisting of the TERT promoter region and the TP53 coding sequence (CDS) with BBAP.15 Somatic mutations were identified by calculating the nucleotide frequencies according to assembly results and comparison with endogenous gene reference sequences.
The HBV genotype of a sequenced sample was determined by assembling its HBV genomic sequence from its NGS data sets using BBAP and HBV reference sequences. The HBV genotype and sequence variation were determined by comparing the assembled HBV genomic sequences to HBV reference sequences of all available genotypes (A-J). The assigned genotype and variants were analyzed for association with HBV integration or mutation at the TERT promoter region.
Plasmid Constructs
The reporter plasmid wild-type (WT)–TERT–luciferase (Luc) was constructed by inserting –47 to –209 of the TERT promoter fragment in front of the luciferase gene in the promoterless pGL4.10 vector. The G(–124)A-TERT-Luc and G(–146)A-TERT-Luc constructs contained the G to A mutation at the –124 and –146 position (relative to ATG) in WT-TERT-Luc. These constructs were kindly provided by Professor Costello at the University of California, San Francisco.16 The HBV-TERT-Luc reporter plasmids were constructed by insertion of the DNA fragments PCR-amplified from HCC of patients M15, M33, M52, M57, and F36, which contained HBV integrations within the TERT promoter region, into the NheI/SacI and HindIII restriction enzyme sites of the pGL4.10 vector. The PCR products amplified by the same primer sets with the DNA from normal liver were constructed as control plasmids in the transfection analysis. The primer sets used for the PCR are listed in Supporting Table S2, and the inserted sequence was confirmed by Sanger sequence analysis.
The details for the plasmids expressing the androgen receptor (pSG5-AR), estrogen receptor alpha (pCMV5-ESR1), and the reporter constructs of 5XARE-Luc and estrogen response element (ERE)–E1b-Luc have been described.9, 17
Results
Validation of the Capture-NGS Platform for Identifying HBV Integration in HCC: Results Comparable to Genomic Sequencing
To examine whether the HBV integration patterns in HCC show any gender differences, the integration sites were identified and compared between male and female patients with HCC. HCCs from 62 male and 39 female patients with HBV-related HCC were included in our analysis, with the clinical information summarized in Supporting Table S1.
Instead of genomic sequencing, we developed a capture-NGS platform to identify the viral integration sites in these HCCs following the flowchart outlined in Fig. 1A. In brief, the tumor gDNA was extracted for library construction, in which the DNA fragments containing HBV were enriched by hybridization with the probes covering the whole HBV genome. In addition to the pure HBV DNA fragments, the DNA fragments containing the HBV-host gene junctions at viral integration sites, namely, the vh-chimera DNA, were enriched by hybridization. Moreover, we included the probes covering the TERT promoter and CDS of TP53 to analyze the two common somatic mutations in the HCC samples. The captured DNA fragments were then processed for NGS and bioinformatics analysis as summarized in Fig. 1A.
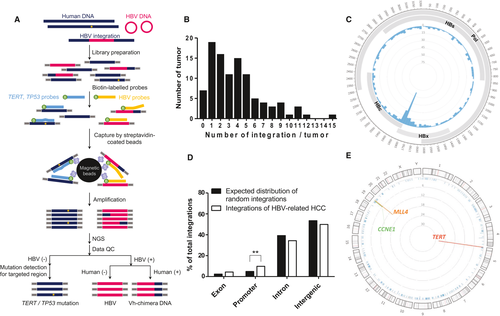
HBV integrations were identified in 94 out of the 101 HCCs in our platform (93.1% of samples containing HBV integration). The number of integrations in these HCCs is summarized in Fig. 1B, ranging from 1 to 15 integrations in individual HCCs. All of the major HBV integrations, including those close to the hotspot genes of TERT, MLL4, and CCNE1 and some others, have been confirmed by junction-specific PCR, with the representative results shown in Supporting Fig. S1. Each chimera DNA was only detected in one HCC as a unique signature for the individual HCC. The integration breakpoints in HBV genome are schematically summarized in Fig. 1C, showing hotspot clustering approximately in the HBV DRI and DRII regions, consistent with previous studies.10 The distribution of HBV integration sites in the HCC genome, exon, promoter, intragenic (intron), and intergenic regions is summarized in Fig. 1D. The integration sites close to the specific cellular genes were plotted (Fig. 1E), with details summarized in Supporting Table S3. The results showed that the integrations located across all of the chromosomes but with hotspots adjacent to the TERT, MLL4, and CCNE1 genes in 28, 14, and 6 out of 101 HCC cases (Fig. 1E). The average number of HBV integrations for one HCC (3.8 integrations/HCC) and the integration patterns were largely consistent with those identified by previous whole-genome sequencing analysis,10 thus validating our capture-NGS platform for identifying HBV integrations in the human HCC genome.
In addition to the HBV integrations, point mutations in the TERT promoter and the CDS of the TP53 gene were also detected using our capture-NGS analysis. Twenty out of 101 HCCs contained TERT promoter mutations (19.8%), and 38 out of 101 HCCs contained TP53 mutations (37.6%). These somatic mutations were validated by Sanger sequencing analysis and summarized in Supporting Table S4. Of note, the TERT promoter mutation and HBV integrations in the TERT promoter region occurred in a mutually exclusive manner, accounting for 47.5% (48 out of 101) of the HCC cases in combination.
HBV Integrations and Point Mutations in the Tert Promoter Region Preferentially Occur in Male Patients with HCC
To examine any gender differences in HBV integration patterns in these HCC cases, the NGS results were stratified by gender. No significant difference was identified between male and female patients with HCC regarding the integration frequency, the integration number/tumor (Fig. 2A), or the breakpoints in the HBV genome (Fig. 2B). For the distribution pattern of integrations in exon/promoter/intron/intergenic regions in the host genome, we observed a significantly higher frequency of HBV integrated in gene promoter regions in male patients with HCC (Fig. 2C). Interestingly, a markedly higher frequency of integrations was observed in the TERT promoter region in male patients with HCC (Figs. 2D and 3) (22/58 male patients with HCC, 6/36 female patients with HCC, P = 0.0285). Such a gender difference, however, did not exist for integrations in the MLL4 and CCNE1 genes (Figs. 2D and 3).
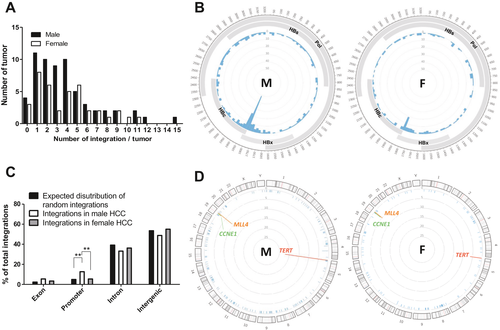
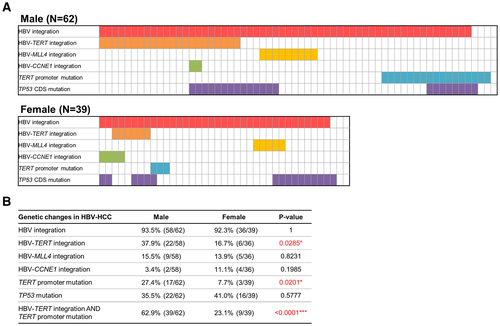
The details for the HBV integrations close to the TERT, MLL4, and CCNE1 genes and the point mutations in the TERT promoter and TP53 CDS regions for these 101 HCC cases are schematically summarized in Fig. 3A (upper panel, male patients with HCC; lower panel, female patients with HCC). Notably, not only the HBV integrations in the TERT promoter region but also the point mutation in the TERT promoter, at –124 relative to the ATG (–124G>A), occurred more frequently in male patients with HCC compared to female patients with HCC (17/62 male, 3/39 female, P = 0.0201). In contrast, no gender difference was found for mutations in the TP53 CDS (22/62 male, 16/39 female, P = 0.5777) (Fig. 3B). As these two types of genetic aberrations at the TERT promoter occur in a mutually exclusive manner, comparison of these two types of genetic changes in combination between male versus female HCCs showed a marked gender difference, which accounted for 39/62 (62.9%) male patients with HCC but only for 9/39 (23.1%) female patients with HCC (P < 0.0001) (Fig. 3B).
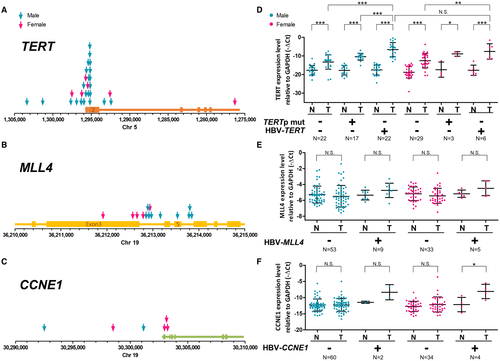
The position of the HBV integration sites in the TERT, MLL4, and CCNE1 gene regions are illustrated in Fig. 4A-C. The integrations were clustered in the promoter region of TERT and CCNE1 but clustered at introns 3-5 of MLL4, which was also consistent with the pattern identified in a previous NGS analysis.10 Moreover, we examined the mRNA expression of TERT (stratified by the promoter integration and promoter point mutation), MLL4, and CCNE1 in male and female HCCs by quantitative RT-PCR analysis. The results indicated that both HBV integrations and point mutations in the TERT promoter region showed elevated TERT expression but that the HBV integrations conferred an even higher TERT expression level than the TERT promoter mutations in male patients with HCC (Fig. 4D). However, the integrations close to introns 3-5 of MLL4 did not increase its expression in HCC (Fig. 4E). For CCNE1, although the frequency was low, we observed that HBV integrations in the promoter region also elevated the gene expression level (Fig. 4F). These results suggested that the HBV integrations adjacent to the TERT and CCNE1 promoter likely activated gene expression but that integration in MLL4 might exert a different effect.
In summary, comparison of the capture-NGS results in male and female patients with HCC indicated that both HBV integration and point mutations in the TERT promoter region, which are associated with higher TERT expression, occurred more commonly in male than in female patients with HCC (P < 0.0001, in combination). Therefore, some gender-specific factors might contribute to the elevation of TERT in male patients with HCC, through either HBV integrations or specific TERT mutations in the TERT promoter region. It may provide a growth advantage for the clonal expansion and carcinogenesis of these hepatocytes, which are thus selected and enriched in the resulting male patients with HCC.
Inverse Effects of Androgen and Estrogen Pathways in Regulating Tert Transcription Through the Integrated HBV in the Tert Promoter Region
We addressed whether the sex hormones affect the transcription of TERT through integrated HBV or point mutations in the TERT promoter using a reporter assay. The reporter plasmid WT-TERT-Luc containing the luciferase gene under the control of the TERT promoter region, covering –209 to –47 relative to ATG, was used to introduce either the HBV integration or point mutations.16 The effects of the androgen and estrogen pathways on the mutant reporter constructs were then evaluated compared with those of the WT construct.
We first focused on the HBV integration type of mutation in the TERT promoter region. In the 28 HCCs with HBV integrations at the TERT promoter region, 27 cases could contain the responsive elements for AR and ERα (Supporting Fig. S2). The responsive elements were highly conserved among most of the integrated HBV, regardless of virus genotype B or C (Supporting Table S5). Among them, a representative HBV-TERT-Luc reporter plasmid containing HBV integrated in the TERT promoter (identified in 1 HCC patient, no. M15) was constructed (Fig. 5A). We observed significantly higher Luc activity in HBV-TERT-Luc(M15) than in WT-TERT-Luc transfected HepG2 cells (~100 times) (Fig. 5B). The results indicated that the integrated HBV induced a strong activation effect on TERT transcription. The AR and ERα expression plasmids were then individually transfected into the cells to examine the effect on WT-TERT-Luc and HBV-TERT-Luc(M15) activity. As control sets, the androgen and estrogen pathways stimulated ARE-Luc and estrogen response element (ERE)–Luc reporter activity in a ligand-dependent manner (Fig. 5C). However, neither pathway had a significant effect on WT-TERT-Luc activity (Fig. 5B), suggesting that the WT-TERT promoter is not under the regulation of sex hormones. For this HBV-TERT-Luc construct, the AR pathway further increased Luc activity in a ligand-dependent manner (Fig. 5B, middle panel). In contrast, the Luc activity of this HBV-TERT-Luc construct was suppressed by the ERα pathway, which was also dependent on the ligand (Fig. 5B, right panel). The results have been validated in another hepatoma cell line, Huh-7 (Supporting Fig. S3). In addition, the HBV-TERT-Luc reporter activity driven by another four HBV integrations at different positions in the TERT promoter region, cloned from HCC of 3 male patients (M33, M52, M57) and 1 female patient (F36), also showed similar hormone responses in both HepG2 and Huh-7 cell lines (Supporting Fig. S4). Finally, a similar result was also confirmed in a female HCC cell line, SNU387 (Supporting Fig. S5). Therefore, the HBV integrations rendered the TERT promoter responsive to sex hormone regulation.
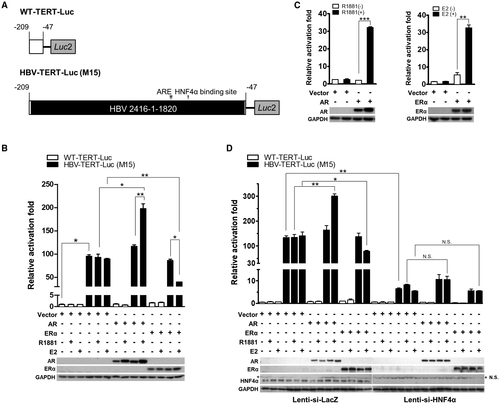
In our previous study, chelation of HNF4α, a major positive transcription factor targeting the core domain of HBV EnhI, was the mechanism responsible for ERα suppression of HBV transcription.9 Thus, we examined the role of HNF4α in these reporter assays. Depletion of HNF4α by lenti-small interfering (si) HNF4α alone dramatically decreased the increment of TERT expression induced by HBV integration (Fig. 5D). Moreover, both the AR and ERα effects on the HBV-TERT-Luc activity were diminished by HNF4α depletion (Fig. 5D). Therefore, HNF4α functions as one fundamental transcription factor responsible for TERT activation through integrated HBV; both androgen-modulating and estrogen-modulating effects are dependent on HNF4α.
The Androgen Pathway Increases Tert Transcription Through the Common Point Mutation in the Tert Promoter, Which is Dependent on GA Binding Protein Transcription Factor Subunit Alpha
Next, we examined whether the TERT promoter mutations –124G>A and –146G>A were also responsive to sex hormone regulation, though the –146G>A mutation was not identified in all 101 HCCs but has been reported in HCC from other cohorts and thus was also included in our analysis.18) The –124G>A and –146G>A mutations were introduced into the WT-TERT-Luc construct, namely, G(–124)A-TERT-Luc and G(–146)A-TERT-Luc, respectively (Fig. 6A). In Huh-7 cells, a significant elevation in the reporter activity in both G(–124)A and G(–146)A-Luc-transfected cells was identified relative to that of the WT-TERT-Luc-transfected cells (Fig. 6B, left panel). The results confirmed the effects of both promoter mutations on TERT transcription in hepatocytes. In the presence of AR, the reporter activity of G(–124)A and G(–146)A-TERT-Luc was further elevated (~3 times) in a ligand-independent manner (Fig. 6B, right panel). In contrast, the reporter activity was not affected by the presence of ERα (Fig. 6C). These findings have been validated in HepG2 cells (Supporting Fig. S6), indicating that the AR, but not the ERα, pathway might also contribute to the up-regulation of the transcription of hepatic TERT through the –124G>A and –146G>A mutations.
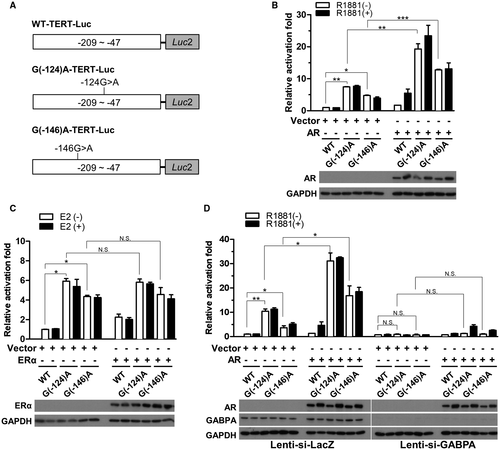
It has been well documented that both –124G>A and –146G>A TERT promoter mutations create new binding sites for GA binding protein transcription factor subunit alpha (GABPA), a key molecule for increasing TERT transcription.16 To evaluate the role of GABPA in mediating AR function to regulate TERT transcription, we depleted GABPA using lenti-si-GABPA and examined the effect on AR function. The results first showed that GABPA knockdown abolished the increase of Luc activity induced by both mutations (Fig. 6D), supporting the fundamental role of GABPA in the activation of the mutated TERT promoter. Interestingly, the increase in Luc activity by AR was also diminished by GABPA depletion (Fig. 6D; Supporting Fig. S6C), indicating that the AR effect on mutant TERT promoter activity was dependent on GABPA. Collectively, the results identified GABPA as the key mediator of AR function to stimulate TERT expression through the common point mutations in the TERT promoter region.
Discussion
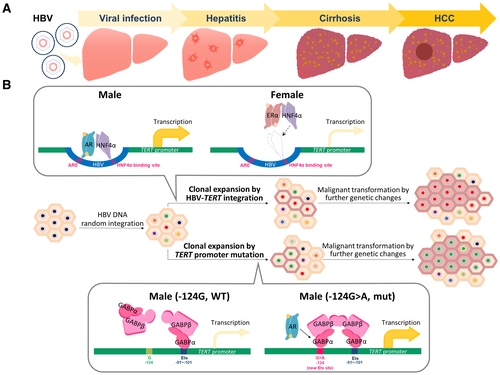
As revealed in a previous NGS analysis, HBV integrations in nontumorous hepatitis liver are distributed randomly over the human genome, without showing any hotspot integration pattern.10, 19 Therefore, the hotspot of the TERT promoter integrations in HCC could be the result of a selection process. Elevation of TERT by HBV integration or somatic nucleotide mutations in the promoter region might provide a growth advantage for the clonal expansion and carcinogenesis of these hepatocytes, which are thus selected for and enriched in the resulting HCC.
According to our capture-NGS analysis, HBV integrations and –124A>G mutations in the TERT promoter region are preferentially enriched in male patients with HCC. In fact, by careful data mining analysis, we found that such a gender difference for the TERT promoter changes also existed in some other NGS analyses. The results showed HBV integrations in 24%-30% of male patients with HCC but only in 12%-17% of female patients with HCC20-22; –124A>G TERT promoter mutation was identified in 32%-33% of male patients with HCC but only in 15%-24% in female patients with HCC.23, 24 Though the sample size of female patients with HCC included in these studies was usually small (fewer than 10 cases), the results did support an enrichment of HBV insertions and TERT promoter mutations in male patients with HCC. Therefore, a mechanism for such genetic changes might exist to induce TERT expression predominantly in hepatocytes from male, but not female, patients.
Our cell culture–based assay indeed supported this possibility, showing that HBV integration and point mutation in the promoter can activate TERT transcription into response to sex hormones (schematically summarized in Fig. 7). The androgen pathway activated, but the ERα pathway suppressed, TERT transcription by targeting the integrated HBV, both of which effects were dependent on HNF4α, a key factor activating HBV transcription25, 26 (Fig. 7B, upper panel). Because the AR and ER responsive sequences are highly conserved among different genotypes of HBV (Supporting Table S5),8, 9 this mechanism could be conserved for different genotypes of HBV integration. In addition, the androgen pathway elevated TERT transcription through TERT promoter mutations at –124G>A mutations, through GABPA binding to the mutated promoter (Fig. 7B, lower panel). Our data thus identified a mechanism to explain a higher elevation of TERT in male hepatocytes through the effects of AR on the integrated HBV and point mutations at the TERT promoter region, though the detailed mechanisms for AR to enhance the HNF4α and GABPA activity needs further investigation.
Why there remained 6 female patients with HCC with HBV-TERT showing elevated TERT mRNA levels comparable with those in the HBV-TERT-containing male patients with HCC needs to be discussed. As an active ERα pathway suppresses the expression of HBV-TERT, it is possible that this mechanism was inactivated in these 6 patients with HCC by a reduction or loss of ERα pathway components, such as by a decrease of estrogen ligand or the ERα protein levels. The estrogen level could be decreased by menopause. The ERα protien in female patients with HCC could be decreased by a mechanism we previously identified, through mutant p53 to elevate the level of microRNA 18a, which targets and decreases the ERα protein in female patients with HCC.27, 28 Accordingly, we found that 4 of these 6 female patients with HCC (F2, F12, F25, and F32) were already at the age of menopause, and mutated TP53 was identified in 3 of them (F2, I162F; F25, I162F; and F32, G245C). Moreover, the quantitative RT-PCR results showed that the mRNA levels for ERα were significantly decreased in three patients with HCC (F16, F32, and F36). All of these events could contribute to reducing the ERα pathway activity, and thus losing the suppression of HBV-TERT expression, explaining the elevated TERT level in these 6 HBV-TERT-containing female patients with HCC.
It has been proposed that TERT elevation is necessary to counteract the exhaustion of hepatocytes in cirrhotic liver, as a prerequisite for subsequent carcinogenesis.29 We thus examined if the HCCs with increased TERT by HBV integration or –124G>A mutation at the TERT promoter is associated with liver cirrhosis. Interestingly, the results showed that the HCCs with TERT elevated by these two genetic changes are significantly associated with liver cirrhosis (47.9% versus 22.6%, P = 0.0077). This suggested that activation of TERT through both genetic changes might be critical for the HCC developed from the cirrhotic livers.
According to this mechanism identified for the integration of HBV in the TERT promoter region, it remains unclear why the male preference pattern was not found in another hotspot gene, MLL4. In contrast to TERT, we found that the HBV integrations were mainly located in introns 3-5 rather than in the promoter region of MLL4. As we found that MLL4 expression was not significantly elevated in HCCs with nearby HBV integrations, some other mechanism for HBV integration to affect MLL4, for example, production of truncated MLL4 protein, is worth examining. The case of CCNE1 is less clear because the case numbers for the HBV integrations were too few to make a generalized statement and, thus, a larger-scale study is needed. Interestingly, we noted that HBV-related HCC showed a mutually exclusive pattern according to the HBV integration at the TERT promoter, mutation at the TERT promoter, and integration at the MLL4 gene, though the reason for that remains unclear. The possibility that MLL4 also functions in stimulating TERT expression is worth testing.
The two genetic events in the TERT promoter regions accounted for about two thirds (39/62, ~63%) of male patients with HCC; however, we noted that TERT expression was still elevated in the other third of male patients with HCC in the absence of either genetic change, though with lower elevation levels (Fig. 4D). Furthermore, TERT was also elevated in most female patients with HCC, although the two types of promoter genetic changes were rare. Therefore, some mechanism other than genetic changes might exist for the elevation of TERT in HCC without promoter genetic changes. For example, several mechanisms have been proposed to overcome TERT silencing, through either specific signaling pathways or specific epigenetic regulations targeting its promoter regulation.30, 31 As a ribonucleoprotein that catalyzes the addition of telomeric repeats to the ends of chromosomes, elevation of TERT activity is critical for the immortality of liver cancer cells. Regardless of the mechanism, TERT is elevated in most HCCs, which, however, is silent in normal hepatocytes. Therefore, TERT overexpression could be a potential therapeutic target for HCC.
Although there are several telomerase inhibitors under development, such as imetelstat and some typical G-quadruplex stabilizers,32, 33 they have not yet been applied to HCC. The molecular mechanisms delineated in the current study might help identify the proper targets to block the elevation of TERT in a specific subgroup of HCC. For example, based on our study, targeting the mechanism by which hepatic AR stimulates TERT expression could be a feasible strategy for blocking the elevation of TERT in male patients with HCC derived from hepatocytes with specific genetic changes in the TERT promoter region.
Potential conflict of interest
Nothing to report.
Acknowledgment
The authors thank the Taiwan Liver Cancer Network (TLCN) for providing samples and related data (all anonymous) for our research.




