Thwart your destiny; effect of nonacoholic fatty liver disease genes on steatosis, liver injury and cirrhosis varies by body mass index
Potential conflict of interest: Nothing to report.
E.K.S. is supported by NIH grants K23 DK080145, RO1 DK106621, and RO1 DK107904, The University of Michigan Biological Sciences Scholars Program, and the University of Michigan Department of Internal Medicine.
Most common human diseases develop because of a combination of endogenous risk combined with exposures. Said another way, these diseases are attributed to genetic risk combined with environmental triggers. Nonalcoholic fatty liver disease (NAFLD) is no exception. There have been both genetic and environmental variables that have been associated with NAFLD. One recent article, however, goes beyond these associations to understand how environmental exposures in the setting of genetic risk variants can exacerbate the development of NAFLD.
Genetic associations studies usually assume an additive genetic model to assess for association with disease. This translates to a model where having 0, 1, or 2 alleles of a genetic variant increases the risk of the disease linearly. However, there are situations where those same alleles instead of increasing risk additively increase risk multiplicatively. That is, having 0, 1, or 2 alleles increases risk under one set of conditions (body mass index [BMI] <25 kg/m2) from 1 to 2 to 4 and another set of conditions (BMI >35 kg/m2) from 1 to 8 to 16 (Fig. 1A). The condition under which the effects of the same genotype changes is called a modifier. Identifying environmental modifiers of genetic risk can be done by looking for gene environment interactions. It is important to identify modifiers of disease because if the modifier can be changed, the increased genetic risk for development of disease can be preferentially and dramatically mitigated.
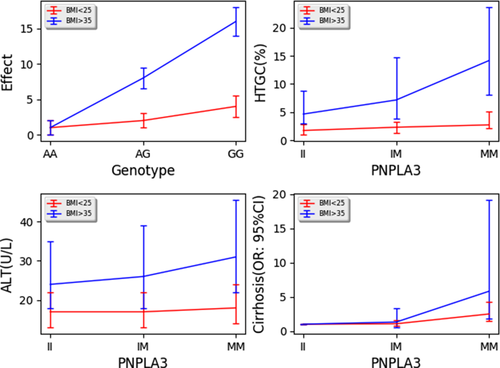
A recent article by Stender et al.1 examined the effect of adiposity on the risk of developing NAFLD in individuals that carried genetic variants encoding patatin-like phospholipase domain containing 3 (PNPLA3) p.I148M (isoleucine to methionine at position 148), TM6SF2 (transmembrane 6 superfamily member 2) p.E167K (glutamic acid to lysine at position 167), and GCKR (glucokinase regulator) p.P446L (proline to leucine). Associations of these variants with hepatic steatosis, nonalcoholic steatohepatitis, and fibrosis has been reported in many cohorts and ancestries.2-5 This article examined whether the effects of these variants on development of NAFLD were modified by BMI. They showed that as BMI increases from less than 25 kg/m2 to 25-30 kg/m2 to 30-35 kg/m2 and to >35 kg/m2, the effect of all three genes on hepatic triglyceride content percent as measured using magnetic resonance spectroscopy in the Dallas Heart Study also increased. The difference in hepatic triglyceride percent median increased from 0.98% in the low-risk PNPLA3-II and PNPLA3-MM, BMI <25 kg/m2 individuals to up to 9.4% in PNPLA3-II and PNPLA3-MM, BMI >35 kg/m2 individuals (Fig. 1B for PNPLA3). Similar interactions were noted by another group using visceral adipose tissue and computed tomography measured fatty liver.6 They repeated analyses using alanine aminotransferase (ALT) as the outcome to test whether it also affected hepatic injury. They found a significant interaction for PNPLA3-M with serum ALT levels where the difference in ALT between PNPLA3-II and PNPLA3-MM, BMI <25 kg/m2 individuals was 1.0 IU/L up to 7 IU/L between PNPLA3-II and PNPLA3-MM, BMI >35 kg/m2 individuals in the Copenhagen Cohort (Fig. 1C). These data corroborate a similar result reported for PNPLA3-M with liver function tests in an Italian cohort.7 Stender et al. also noted that the difference in odds ratio (OR) between PNPLA3-II and PNPLA3-MM, BMI <25 kg/m2 individuals of 1.5-4.8 between PNPLA3-II and PNPLA3-MM, BMI >35 kg/m2 individuals suggesting an interaction with this liver disease measure also (Fig. 1D). They observed statistically significant, but smaller, interactions for the GCKR and TM6SF2 variants in affecting hepatic triglyceride content, but not with ALT and cirrhosis. This may be attributed to biology or to low power to detect an effect for these variants for ALT and cirrhosis.
Even though the percentage difference in liver fat and ALT in Stender et al. across obesity groups seems small, these estimates come from cross-sectional measures that can sum to larger effects over a lifetime. Although BMI is also genetically influenced,8 it is modifiable. Therefore, by decreasing the modifier, in this case BMI, strides may be made toward preventing common genetic diseases in those genetically susceptible. From these population-based data, it is estimated that PNPLA3 MM homozygotes may be able to reduce their hepatic triglyceride content from 14% to 3%, their ALT from 31 to 18 IU/L and their odds of developing cirrhosis from 5.8 to 2.5 by reducing their BMI from >35 to <25 kg/m2. In contrast, PNPLA3 II individuals will only reduce their hepatic triglyceride content from 4.7% to 1.8% and their ALT from 24 to 11 IU/L by reducing their BMI from >35 to < 25 kg/m2. The OR for cirrhosis are standardized to 1.0 for PNPLA3 II individuals, so the comparison is not possible. Hence, weight loss may be preferentially beneficial for individuals at genetic risk of developing disease, for example, the ones that carry PNPLA3 M alleles. By identifying these environmental modifiers of genetic disease risk, we can better counsel people with genetic risk of developing human disease to mitigate all stages of disease from steatosis, to liver injury, to cirrhosis. Furthermore, because of the mathematical nature of a gene environment interaction, individuals at risk can improve their outcome more than individuals not at genetic risk of disease by reducing the modifier.
As proof of concept, two small studies have shown that individuals with the PNPLA3 NAFLD risk allele (M) lose more liver fat after weight loss than individuals carrying the wild-type allele (I). One set of investigators placed eight PNPLA3 MM homozygotes and 10 PNPLA3 II homozygotes matched by age, sex, body mass index, and liver fat on a hypocaloric low-carbohydrate diet for 6 days and measured liver fat measured by proton magnetic resonance spectroscopy before and after the diet.9 Both groups lost, on average, 3.1 ± 0.5 kg of body weight. However liver fat decreased by 45% in the PNPLA3 MM compared to 18% in the PNPLA3 II group. In a second study, 84 obese individuals were followed after bariatric surgery for weight loss and for liver fat using magnetic resonance imaging.10 Individuals that were PNPLA3 MM lost more weight and liver fat 1 year after surgery compared to PNPLA3 II individuals. These studies reinforce the idea that decreasing exposure to a disease modifier can mitigate genetic risk of disease and provide preferential benefit to individuals at genetic risk of developing disease. Therefore, genetic risk factors of disease as well as of environmental variables that can modify risk are important to know given that they can differentially influence the risk versus benefit of interventions for disease. Future larger clinical trials by genotype will be needed to confirm the benefits of weight loss for individuals with genetic risk and can be used to guide broader precision medicine approaches to common genetically influenced diseases.
-
Elizabeth K. Speliotes, M.D., Ph.D., M.P.H.
-
Divisions of Gastroenterology
-
Department of Internal Medicine
-
Department of Computational Medicine and Bioinformatics
-
University of Michigan
-
Ann Arbor, MI
REFERENCES
Author names in bold designate shared co-first authorship.




