Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study
Potential conflict of interest: Dr. Schork consults and owns stock in Human Longevity. He is employed and owns stock in MYI and CEGC. Dr. Sirlin consults for Fibrogen, Tobira, and Virtualscopic.
Supported in part by the American Gastroenterological Association Foundation-Sucampo-ASP Designated Research Award in Geriatric Gastroenterology (to R.L.) and a T. Franklin Williams Scholarship Award (to R.L.) and by Atlantic Philanthropies Inc., the John A. Hartford Foundation, OM, the Association of Specialty Professors, and the American Gastroenterological Association (grant K23-DK090303). Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abstract
Nonalcoholic fatty liver disease is associated with metabolic risk factors including hypertension and dyslipidemia and may progress to liver fibrosis. Studies have shown that hepatic steatosis and fibrosis are heritable, but whether they have a significant shared gene effect is unknown. This study examined the shared gene effects between hepatic steatosis and fibrosis and their associations with metabolic risk factors. This was a cross-sectional analysis of a prospective cohort of well-characterized, community-dwelling twins (45 monozygotic, 20 dizygotic twin pairs, 130 total subjects) from southern California. Hepatic steatosis was assessed with magnetic resonance imaging-proton density fat fraction and hepatic fibrosis with magnetic resonance elastography. A standard bivariate twin additive genetics and unique environment effects model was used to estimate the proportion of phenotypic variance between two phenotypes accounted for by additive genetic effects and individual-specific environmental effects. Genetic correlations estimated from this model represent the degree to which the genetic determinants of two phenotypes overlap. Mean (± standard deviation) age and body mass index were 47.1 (±21.9) years and 26.2 (±5.8) kg/m2, respectively. Among the cohort, 20% (26/130) had hepatic steatosis (magnetic resonance imaging-proton density fat fraction ≥5%), and 8.2% (10/122) had hepatic fibrosis (magnetic resonance elastography ≥3 kPa). Blood pressure (systolic and diastolic), triglycerides, glucose, homeostatic model assessment of insulin resistance, insulin, hemoglobin A1c, and low high-density lipoprotein had significant shared gene effects with hepatic steatosis. Triglycerides, glucose, homeostatic model assessment of insulin resistance, insulin, hemoglobin A1c, and low high-density lipoprotein had significant shared gene effects with hepatic fibrosis. Hepatic steatosis and fibrosis had a highly significant shared gene effect of 0.756 (95% confidence interval 0.716-1, P < 0.0001). Conclusions: Genes involved with steatosis pathogenesis may also be involved with fibrosis pathogenesis. (Hepatology 2016;64:1547-1558)
Abbreviations
-
- AE model
-
- model including additive genetics and environmental effects
-
- ALT
-
- alanine aminotransferase
-
- BMI
-
- body mass index
-
- CI
-
- confidence interval
-
- DBP
-
- diastolic blood pressure
-
- HbA1c
-
- hemoglobin A1c
-
- HDL
-
- high-density lipoprotein
-
- HOMA-IR
-
- homeostatic model assessment of insulin resistance
-
- LDL
-
- low-density lipoprotein
-
- MRE
-
- magnetic resonance elastography
-
- MRI-PDFF
-
- magnetic resonance imaging-proton density fat fraction
-
- NAFLD
-
- nonalcoholic fatty liver disease
-
- NASH
-
- nonalcoholic steatohepatitis
-
- rE
-
- shared environmental determination
-
- rG
-
- shared genetic determination
-
- SBP
-
- systolic blood pressure
-
- UCSD
-
- University of California at San Diego
Nonalcoholic fatty liver disease (NAFLD) is comprised of a spectrum of liver pathologies characterized by hepatic steatosis in patients with little to no history of alcohol use or secondary causes of hepatic steatosis.1 Nonalcoholic steatohepatitis (NASH) is an advanced form of NAFLD and predisposes patients to the development of hepatic fibrosis, which is associated with increased risks of cirrhosis, mortality, and liver transplantation.2-5 NAFLD, including its complications, is now a leading cause of liver disease in the United States and worldwide.6-8 Due to the heavy disease burden of NAFLD and its associated morbidity and mortality, there is a great need to characterize the heritability of NAFLD to identify patients who may be at risk for the disease, improve the understanding of NAFLD pathogenesis, and identify potential targets for treatment.
Hepatic steatosis represents the initial step for the pathogenesis of NASH and hepatic fibrosis, and we have previously demonstrated in twin models that both hepatic steatosis and fibrosis are heritable traits.9 Various genes, including PNPLA-3 and TM6SF2, are associated with the development of hepatic steatosis and fibrosis, although variations in these genes do not account for all the variance seen in hepatic steatosis and fibrosis, and additional genes remain to be identified.10-15 But while hepatic steatosis can progress to hepatic fibrosis, it is unknown if there are direct genetic links between these two traits. Hepatic fibrosis has been shown to be the most important predictor of mortality and liver transplantation in NAFLD patients,5, 16 and an improved understanding of the shared heritability between hepatic steatosis and fibrosis may elucidate potential targets for NAFLD prevention and treatment. If steatosis and fibrosis gene regulation significantly overlap, then it is plausible that improvement in steatosis by a shared mechanistic pathway may eventually trigger improvement in fibrosis in the context of targeting specific nodal points in the mechanistic pathway. Additionally, studies have shown NAFLD to be associated with metabolic risk factors including obesity, hypertension, dyslipidemia, and insulin resistance17-20, and there is a genetic component to this association.21 However, further studies are needed to characterize the genetic association between hepatic steatosis, fibrosis, and individual metabolic risk factors.
Using a prospective study design of community-dwelling monozygotic and dizygotic twins, we evaluated if study participants with genetic susceptibilities to hepatic steatosis also have genetic susceptibilities to hepatic fibrosis. We also evaluated the genetic susceptibilities of hepatic steatosis and fibrosis with metabolic risk factors, including blood pressure, cholesterol levels, triglycerides, insulin resistance, and diabetes. Mathematical models involving additive genetics and unique environmental effects (called AE models) were constructed for the cohort to distinguish between the shared genetic and environmental determination of individual traits. Magnetic resonance imaging-proton density fat fraction (MRI-PDFF) and magnetic resonance elastography (MRE), two novel, accurate, and noninvasive imaging biomarkers, were respectively used to assess for hepatic steatosis and fibrosis in this prospective study.
Participants and Methods
EXPERIMENTAL DESIGN
This was a cross-sectional analysis of a prospectively recruited cohort of monozygotic and dizygotic twin pairs living in southern California. All twin pairs underwent clinical research assessments, including medical history, physical and anthropometric exams, and biochemical testing, at the University of California at San Diego (UCSD) NAFLD Research Center.15, 21-23 Participants also underwent MRI-PDFF for hepatic steatosis and MRE for hepatic fibrosis at the UCSD MR3T Research Laboratory. Clinical and imaging visits were performed on the same day for each twin pair, and the study took place from 2012 to 2015. All participants provided written informed consent before enrolling in the study. The study protocol was approved by the UCSD institutional review board.
INCLUSION CRITERIA
Participants were included in the study if they were twins at least 18 years old who provided written informed consent. The zygosity of the majority of twin pairs as monozygotic or dizygotic had been confirmed through genetic testing before the participants enrolled in the study. A previously published questionnaire, as described by Boyd et al.,24 was used to further confirm twinship status (see Supporting Information for details).
EXCLUSION CRITERIA
Participants were excluded from the study if they met any of the following criteria: (1) significant alcohol intake (>10 g/day in females or >20 g/day in males) for at least 3 consecutive months over the previous 12 months or if the quantity of alcohol consumed could not be reliably ascertained; (2) clinical or biochemical evidence of liver diseases other than NAFLD, including hepatitis B, hepatitis C, alpha-1-antitrypsin deficiency, hemochromatosis, Wilson's disease, autoimmune hepatitis, polycystic liver diseases, cholestatic liver diseases, and vascular liver diseases; (3) chronic illnesses associated with hepatic steatosis, including human immunodeficiency virus infection, type I diabetes mellitus, celiac disease, cystic fibrosis, lipodystrophy, dysbetalipoproteinemia, and glycogen storage diseases; (4) use of drugs known to cause hepatic steatosis, including amiodarone, glucocorticoids, methotrexate, l-asparaginase, and valproic acid for at least 3 out of the previous 6 months; (5) history of bariatric surgery, including roux-en-Y gastric bypass and gastroplasty; (6) presence of systemic infectious illnesses; (7) females who were pregnant or nursing at the time of the study; (8) contraindications to MRI, including metal implants, claustrophobia, and body circumference greater than that of the imaging chamber; (9) any other condition(s) which, based on the principal investigator's opinion, may significantly affect the participant's compliance, competence, or ability to complete the study.
DEFINITION OF NAFLD
Participants were considered to have NAFLD if they had hepatic steatosis (MRI-PDFF ≥5%) and no secondary causes of hepatic steatosis due to factors including the use of steatogenic medications, other liver diseases, and significant alcohol intake (see Exclusion Criteria above for details).
CLINICAL RESEARCH ASSESSMENT
All participants underwent clinical research assessments at the UCSD NAFLD Research Center (see Supporting Information for details).
GENOTYPING
DNA samples were extracted from whole blood samples collected during the clinical research visit. Genotyping was performed by Human Longevity Inc. (San Diego, CA).
PRIMARY OUTCOME
The primary outcome was the presence of shared gene effect between hepatic steatosis and hepatic fibrosis.
SECONDARY OUTCOMES
The secondary outcomes were the shared gene of hepatic steatosis and fibrosis with the following metabolic risk factors: systolic blood pressure (SBP), diastolic blood pressure (DBP), total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides, ferritin, glucose, homeostatic model assessment of insulin resistance (HOMA-IR), insulin, and hemoglobin A1c (HbA1c).
MRI
MRI was performed at the UCSD MR3T Research Laboratory using the 3T research scanner (GE Signa EXCITE HDxt; GE Healthcare, Waukesha, WI) with all participants in the supine position, on the same day as the clinical research visit, to reduce potential confounding factors. MRI-PDFF was used to measure hepatic steatosis, and MRE was used to measure hepatic fibrosis. MRI-PDFF has been shown to be a highly precise, accurate, and reproducible noninvasive biomarker to quantify hepatic fat content,25, 26 correlates well with magnetic resonance spectroscopy (r2 = 0.99, P < 0.001),22, 23 and is superior to noninvasive imaging techniques such as ultrasound and computed tomography for measuring hepatic fat content,27 even in iron-overloaded livers that may coexist with NAFLD livers.28 MRI-PDFF has also been shown to correlate well with histology from contemporaneous liver biopsies.29, 30 MRE has been shown to be a highly accurate, noninvasive biomarker to estimate hepatic fibrosis quantified by liver stiffness values in units of kilopascals31 and has been shown to be more accurate than clinical prediction rules32 and ultrasound-based acoustic radiation force impulse imaging33 for quantifying hepatic fibrosis. Please see Supporting Information for a description of the magnetic resonance procedures.
JUSTIFICATION FOR NOT USING LIVER BIOPSY TO ASSESS FOR HEPATIC STEATOSIS AND FIBROSIS
Due to the invasive nature of liver biopsies, it would be unethical to perform liver biopsies in study participants with no clinical indications for liver biopsies.1 Therefore, we used noninvasive imaging techniques to quantify hepatic steatosis and fibrosis. MRI-PDFF has been shown to be accurate for estimating hepatic steatosis and more precise than liver biopsies.25 MRE has also been shown to be accurate for estimating hepatic fibrosis.31-33
STATISTICAL ANALYSIS
Patients' demographic, anthropometric, clinical, and biochemical characteristics were summarized. Categorical variables were shown as counts and percentages, and associations were tested using a chi-squared test or Fisher's exact test. Normally distributed continuous variables were shown as mean (± standard deviation), and differences between groups were analyzed using a two-independent samples t test or Wilcoxon-Mann-Whitney test. Odds ratios were derived from generalized estimating equations (PROC GENMOD) to account for intrapair correlations within twinships. A two-tailed P < 0.05 was considered statistically significant. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC).
AE models were used to estimate the shared genetic determination (rG) and shared environmental determination (rE) between twin pairs. In the classical twin study of sets of monozygotic and dizygotic twins, four latent factors can account for the variance of any phenotype: additive genetic effects (A); nonadditive genetic effects, including dominance (D); common or shared environmental effects (C); and nonshared or individual-specific environmental effects (E).34 Because monozygotic twins are presumed to be genetically identical, they correlate perfectly (r = 1.0) with respect to both additive and nonadditive genetic effects. Dizygotic twins share, on average, 50% of their genes, resulting in correlations of 0.50 for additive genetic effects and 0.25 for nonadditive genetic effects. The C term is defined as environmental factors that make twins similar; hence, common environmental factors correlate 1.0 across twin pairs, regardless of zygosity. The E term represents environmental factors that lead to differences between twins. Because these are individual-specific factors, they are assumed to be uncorrelated across twins. Error is assumed to be random across individuals, so measurement error forms part of the estimate of E in these analyses. These latent factors comprise what are referred to as the univariate ACE or ADE models; due to model underidentification, an ACDE model cannot be tested in the classical twin design.34
The ACE and ADE models are easily extended to the multivariate case.34 In addition to genetic and environmental sources of variance, sources of covariance can be examined in the bivariate model. In the present study, we used bivariate models to compute genetic correlations between two phenotypes. A phenotypic correlation measures shared variance; a genetic correlation measures shared genetic variance. More specifically, a phenotypic correlation is defined as the total covariance (genetic plus environmental) of two variables divided by the square root of the product of the total variance of variable 1 and the total variance of variable 2. After decomposing the sources of variance in the bivariate model, we computed genetic correlations. These are defined as the genetic covariance divided by the square root of the product of the genetic variance of variable 1 and the genetic variance of variable 2. The analyses were performed using OpenMx, a structural equation modeling software package for genetically informative data (http://openmx.psyc.virginia.edu). Prior to the model fitting, the measures were adjusted for controlling age, gender, and ethnicity. Overall, AE models tended to provide the best fits to the data. Consequently, the genetic effects estimated in these AE models refer to broad-sense heritability, reflecting the proportion of phenotypic variance accounted for by the combined effect of all genetic influences (A+D).
SAMPLE SIZE ESTIMATION
In previous studies, the heritability of hepatic steatosis ranged from 0.37, when hepatic steatosis was assessed using ultrasound and serum alanine aminotransferase (ALT) levels,35 to almost 1.0, when hepatic steatosis was assessed using MRI in obese Hispanic probands and their relatives.36 We have also previously estimated the heritability of hepatic steatosis and fibrosis to be approximately 0.5.9 Based on these numbers, we anticipated that the heritability of hepatic steatosis and fibrosis with one another should also be approximately 0.5. It has been shown that, to detect an additive genetic component of 0.4-0.8 in an ACE model, approximately 36-74 twin pairs are needed to produce a power of 0.95 with an alpha value of 0.05.37 Therefore, the 65 twin pairs in this study should provide an adequate sample size to assess the heritability of steatosis and fibrosis in our population.
Results
BASELINE CHARACTERISTICS
Included in the study were 130 participants (45 monozygotic twin pairs, 20 dizygotic twin pairs) who underwent clinical research assessments and imaging with MRI-PDFF and MRE. Initially, 438 participants were assessed for eligibility, 152 provided informed consent, and 130 were included in the final analysis (see Supporting Fig. S1 for details). The mean (± standard deviation) age was 47.1 (±21.9) years, and the mean (± standard deviation) body mass index (BMI) was 26.2 (±5.8) kg/m2. Among the cohort, 26/130 (20%) had hepatic steatosis (MRI-PDFF ≥5%) and 10/122 (8.2%) had hepatic fibrosis (MRE ≥3 kPa). Compared to twins without NAFLD, twins with NAFLD were significantly older (54.9 ± 17.3 years versus 45.2 ± 22.5 years, P = 0.04) and had a higher BMI (31.5 ± 4.8 kg/m2 versus 24.8 ± 5.1 kg/m2, P < 0.0001). As expected, twins with NAFLD also had significantly higher measurements of hepatic steatosis by MRI-PDFF (10.7 ± 5.1 versus 2.4 ± 0.9, P < 0.0001) and hepatic fibrosis by MRE (3.0 ± 1.2 versus 2.2 ± 0.4, P < 0.0001). Detailed demographic, biochemical, and imaging data of the cohort, stratified by the presence or absence of NAFLD, are summarized in Table 1.
| Twins With | Twins Without | |||
|---|---|---|---|---|
| NAFLD | NAFLD | |||
| (MRI-PDFF | (MRI-PDFF | |||
| Overall | ≥5%) | <5%) | P | |
| Number | 130 | 26 | 104 | |
| Demographics | ||||
| Age (years) | 47.1 (21.9) | 54.9 (17.3) | 45.2 (22.5) | 0.0417 |
| Sex (% male) | 35 (26.9%) | 11 (42.3%) | 24 (23.1%) | 0.0480 |
| Race | 0.0510 | |||
| White | 104 (80.0%) | 18 (69.2%) | 86 (82.7%) | |
| Hispanic | 18 (13.9%) | 5 (19.2%) | 13 (12.5%) | |
| Asian | 6 (4.6%) | 1 (3.9%) | 5 (4.8%) | |
| Hawaiian/Pacific Islander | 2 (1.5%) | 2 (7.7%) | 0 (0%) | |
| Physical | ||||
| Height (cm) | 165.8 (8.2) | 167.0 (10.7) | 165.4 (7.5) | 0.4981 |
| Weight (kg) | 72.1 (17.9) | 88.8 (20.5) | 67.8 (14.3) | <0.0001 |
| BMI (kg/m2) | 26.2 (5.8) | 31.5 (4.8) | 24.8 (5.1) | <0.0001 |
| SBP (mm Hg) | 126.0 (19.6) | 135.5 (16.7) | 123.6 (19.6) | 0.0052 |
| DBP (mm Hg) | 78.7 (12.4) | 82.9 (13.0) | 77.6 (12.1) | 0.0494 |
| Waist circumference (cm) | 88.8 (12.9) | 100.0 (10.6) | 85.9 (11.9) | <0.0001 |
| Hip circumference (cm) | 99.6 (11.3) | 107.5 (11.6) | 97.7 (10.4) | <0.0001 |
| Laboratory data | ||||
| Glucose (mg/dL) | 89.3 (18.5) | 101.6 (34.3) | 86.2 (9.5) | 0.0013 |
| Insulin (U/L) | 8.4 (5.5) | 12.6 (7.1) | 7.4 (4.5) | 0.0005 |
| HbA1c | 5.8 (0.5) | 6.1 (0.7) | 5.7 (0.3) | 0.0011 |
| HOMA-IR | 1.9 (1.4) | 3.1 (1.9) | 1.6 (1.1) | 0.0001 |
| AST (U/L) | 23.6 (9.9) | 27.0 (16.6) | 22.7 (7.2) | 0.1676 |
| ALT (U/L) | 22.9 (18.1) | 35.0 (32.8) | 19.8 (10.0) | 0.0008 |
| Alkaline phosphatase (U/L) | 69.1 (19.6) | 69.7 (15.1) | 69.0 (20.6) | 0.6589 |
| Total bilirubin (mg/dL) | 0.5 (0.3) | 0.5 (0.4) | 0.5 (0.2) | 0.7893 |
| Direct bilirubin (mg/dL) | 0.1 (0.1) | 0.1 (0.1) | 0.1 (0.1) | 0.4888 |
| Albumin (g/dL) | 4.5 (0.3) | 4.5 (0.2) | 4.5 (0.4) | 0.3867 |
| GGT (U/L) | 23.2 (19.9) | 33.1 (29.6) | 20.7 (15.7) | 0.0031 |
| Total cholesterol (mg/dL) | 196.0 (40.8) | 200.6 (36.1) | 194.8 (42.1) | 0.3330 |
| HDL-cholesterol (mg/dL) | 66.4 (21.7) | 51.0 (15.9) | 70.4 (21.2) | <0.0001 |
| LDL-cholesterol (mg/dL) | 111.0 (34.7) | 119.8 (30.7) | 108.8 (35.5) | 0.0489 |
| Triglycerides (mg/dL) | 92.8 (53.3) | 150.7 (71.1) | 78.0 (35.0) | <0.0001 |
| White blood cell count (×103/μL) | 5.7 (1.3) | 6.4 (1.5) | 5.5 (1.2) | 0.0055 |
| Hemoglobin (g/dL) | 13.7 (1.2) | 14.0 (1.4) | 13.6 (1.2) | 0.1038 |
| Hematocrit (%) | 40.6 (3.3) | 41.5 (3.8) | 40.4 (3.2) | 0.0832 |
| Platelet count ((×103/μL) | 251.7 (51.5) | 253.0 (57.7) | 251.4 (50.2) | 0.8753 |
| INR | 1.1 (0.3) | 1.1 (0.4) | 1.1 (0.3) | 0.6387 |
| Ferritin (ng/mL) | 100.9 (90.7) | 134.4 (144.4) | 92.2 (69.2) | 0.1769 |
| Imaging data | ||||
| MRI-PDFF (%) | 4.034 (4.11) | 10.740 (5.08) | 2.358 (0.87) | <0.0001 |
| MRE (kPa) | 2.327 (0.74) | 3.004 (1.23) | 2.152 (0.40) | <0.0001 |
| PNPLA3 phenotype (n = 87) | 0.8360 | |||
| CC | 39 (44.8%) | 9 (45.0%) | 30 (47.8%) | |
| CG | 38 (43.7%) | 8 (40.0%) | 30 (44.8%) | |
| GG | 10 (11.5%) | 3 (15.0%) | 7 (10.4%) |
- Mean values are provided with standard deviation in parentheses, unless otherwise noted as n (%). Differences between participants with and without NAFLD were evaluated with t tests or the Wilcoxon-Mann-Whitney test for continuous variables and the chi-squared or Fisher exact test for categorical variables.
- Bold indicates significant P values <0.05.
- Abbreviations: AST, aspartate aminotransferase; GGT, gamma-glutamyl transpeptidase; INR, international normalized ratio.
SHARED GENE EFFECT
Using AE models, the shared gene effects (rG) between hepatic steatosis, fibrosis, and metabolic risk factors are summarized below.
Shared Gene Effects Between Hepatic Steatosis and Metabolic Risk Factors
There were significant shared gene effects between hepatic steatosis, as measured by MRI-PDFF, and BMI at 0.534 (95% confidence interval [CI] 0.305-0.713), P = 3.19e-5; SBP at 0.360 (95% CI 0.052-0.636), P = 0.023; DBP at 0.444 (95% CI 0.444-0.742), P = 0.0071; HDL cholesterol at -0.451 (95% CI -0.643 to -0.216), P = 3.57e-4; triglycerides at 0.678 (95% CI 0.585-0.830), P = 4.69e-8; glucose at 0.716 (95% CI 0.716-1), P = 1.64e-4; HOMA-IR at 0.490 (95% CI 0.212-0.739), P = 8.71e-4; insulin at 0.289 (95% CI 0.017-0.531), P = 0.038; and HbA1c at 0.588 (95% CI 0.588-1), P = 9.83e-4. There were no significant shared gene effects between hepatic steatosis and total cholesterol, LDL cholesterol, and ferritin (Table 2). Significant shared gene effects between hepatic steatosis and metabolic risk factors are shown in Fig. 1A.
| Hepatic Steatosis (MRI-PDFF) | Hepatic Fibrosis (MRE) | |||
|---|---|---|---|---|
| Trait | rG Estimate (95% CI) | P | rG Estimate (95% CI) | P |
| BMI | 0.534 (0.305-0.713) | 3.19e-5 | 0.493 (0.493-0.845) | 0.00649 |
| Blood pressure* | ||||
| SBP | 0.360 (0.052-0.636) | 0.023 | 0.308 (-0.137 to 0.688) | 0.160 |
| DBP | 0.444 (0.444-0.742) | 0.0071 | 0.257 (0.257-0.723) | 0.292 |
| Total cholesterol | -0.021 (-0.397 to -0.014) | 0.903 | -0.299 (-0.839 to 0.218) | 0.243 |
| LDL-cholesterol | 0.076 (-0.134 to 0.090) | 0.658 | -0.127 (-0.609 to -0.053) | 0.600 |
| HDL-cholesterol | -0.451 (-0.643 to -0.216) | 3.57e-4 | -0.614 (-0.890 to -0.614) | 5.74e-4 |
| Triglycerides | 0.678 (0.585-0.830) | 4.69e-8 | 0.657 (0.657-1) | 3.44e-4 |
| Ferritin | 0.370 (-1 to 1) | 0.445 | 1 (-1 to 1) | 0.057 |
| Glucose | 0.716 (0.716-1) | 1.64e-4 | 0.746 (0.746-1) | 0.0029 |
| HOMA-IR | 0.490 (0.212-0.739) | 8.71e-4 | 0.610 (0.218-1) | 0.0025 |
| Insulin | 0.289 (0.017-0.531) | 0.038 | 0.429 (0.167-0.735) | 0.023 |
| HbA1c | 0.588 (0.588-1) | 9.83e-4 | 0.566 (0.566-1) | 0.015 |
| Liver fibrosis (MRE) | 0.756 (0.716-1) | 2.54e-5 | N/A | N/A |
- Significant (P < 0.05) coefficients are designated in bold type.
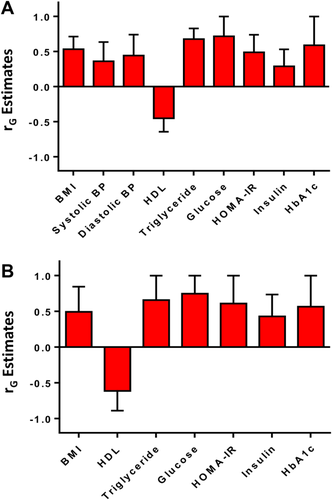
Shared Gene Effects Between Hepatic Fibrosis and Metabolic Risk Factors
There were significant shared gene effects between hepatic fibrosis, as measured by MRE, and BMI at 0.493 (95% CI 0.493-0.845), P = 0.00649; HDL cholesterol at -0.614 (95% CI -0.890 to -0.614), P = 5.74e-4; triglycerides at 0.657 (95% CI 0.657-1), P = 3.44e-4; glucose at 0.746 (95% CI 0.746-1), P = 0.0029; HOMA-IR at 0.610 (95% CI 0.218-1), P = 0.0025; insulin at 0.429 (95% CI 0.167-0.735), P = 0.023; and HbA1c at 0.566 (95% CI 0.566-1), P = 0.015. There were no significant shared gene effects between hepatic fibrosis and SBP, DBP, total cholesterol, LDL cholesterol, and ferritin (Table 2). Significant shared gene effects between hepatic fibrosis and metabolic risk factors are shown in Fig. 1B.
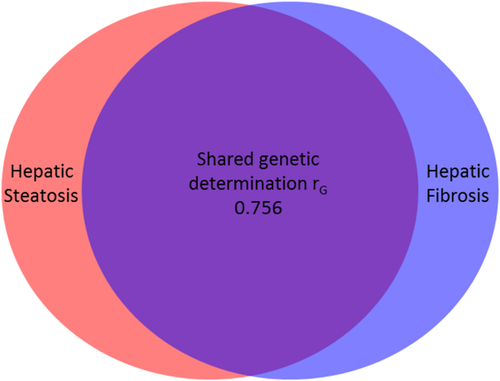
Shared Gene Effect Between Hepatic Steatosis and Fibrosis
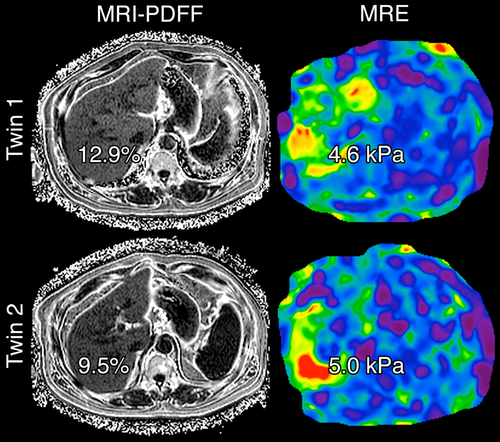
There was a significant shared gene effect between hepatic steatosis and fibrosis at 0.756 (95% CI 0.716-1), P = 2.54e-5 (Table 2 and Fig. 2). Figure 3 depicts MRI-PDFF images and MRE elastograms of a representative pair of 60-year-old male twins with both hepatic steatosis and fibrosis traits.
SHARED ENVIRONMENTAL EFFECT
Using AE models, the shared environmental effects (rE) between hepatic steatosis, fibrosis, and metabolic risk factors are summarized below.
Shared Environmental Effect Between Hepatic Steatosis and Metabolic Risk Factors
There was a significant shared environmental effect between hepatic steatosis, as measured by MRI-PDFF, and ferritin at 0.307 (95% CI 0.019-0.544), P = 0.037. There were no other significant shared environmental effects between hepatic steatosis and other metabolic risk factors (Table 3).
| Hepatic Steatosis (MRI-PDFF) | Hepatic Fibrosis (MRE) | |||
|---|---|---|---|---|
| Trait | rE Estimate (95% CI) | P | rE Estimate (95% CI) | P |
| BMI | 0.087 (-0.188 to 0.352) | 0.0538 | -0.067 (-0.345 to 0.226) | 0.654 |
| Blood pressure* | ||||
| SBP | 0.177 (-0.098 to 0.426) | 0.204 | 0.116 (-0.161 to 0.378) | 0.411 |
| DBP | 0.181 (-0.104 to 0.438) | 0.211 | 0.129 (-0.160 to 0.399) | 0.381 |
| Total cholesterol | 0.178 (-0.128, 0.452) | 0.252 | -0.023 (-0.322 to 0.277) | 0.883 |
| LDL-cholesterol | 0.213 (-0.075 to 0.467) | 0.146 | -0.065 (-0.343 to 0.223) | 0.660 |
| HDL-cholesterol | -0.229 (-0.488 to -0.069) | 0.129 | 0.030 (-0.279 to 0.327) | 0.851 |
| Triglycerides | 0.236 (-0.045 to 0.481) | 0.097 | 0.123 (-0.190 to 0.415) | 0.445 |
| Ferritin | 0.307 (0.019-0.544) | 0.037 | -0.189 (-0.415 to 0.083) | 0.167 |
| Glucose | -0.070 (-0.332 to 0.202) | 0.617 | -0.006 (-0.271 to 0.262) | 0.968 |
| HOMA-IR | 0.015 (-0.271 to 0.298) | 0.917 | -0.166 (-0.430 to 0.125) | 0.261 |
| Insulin | 0.088 (-0.211 to 0.370) | 0.565 | -0.210 (-0.477 to 0.096) | 0.176 |
| HbA1c | 0.200 (-0.098 to 0.460) | 0.185 | 0.089 (-0.195 to 0.360) | 0.543 |
| Liver fibrosis (MRE) | -0.052 (-0.359 to 0.278) | 0.758 | N/A | N/A |
- Significant (P < 0.05) coefficients are designated in bold type.
Shared Environmental Effect Between Hepatic Fibrosis and Metabolic Risk Factors
There were no significant shared environmental effects between hepatic fibrosis, as measured by MRE, and metabolic risk factors (Table 3).
PREDICTORS OF NAFLD IN OVERALL COHORT
In our overall cohort of twins, generalized estimating equations were used to estimate the odds ratios of demographic, anthropometric, and laboratory variables for predicting NAFLD. Significant odds ratios as predictors of NAFLD included weight at 1.07 (95% CI 1.04-1.11), P < 0.0001; BMI at 1.26 (95% CI 1.12-1.43), P = 0.0002; BMI >30 at 6.42 (95% CI 2.40-17.16), P = 0.0002; SBP at 1.03 (95% CI 1.01-1.04), P = 0.0108; waist circumference at 1.10 (95% CI 1.05-1.15), P = 0.0002; hip circumference at 1.09 (95% CI 1.04-1.14), P = 0.0002; glucose at 1.03 (95% CI 1.01-1.05), P = 0.0132; HbA1c at 4.31 (95% CI 1.75-10.65), P = 0.0015; HOMA-IR at 1.63 (95% CI 1.08-2.44), P = 0.0189; ALT at 1.03 (95% CI 1.01-1.06), P = 0.0104; HDL cholesterol at 0.95 (95% CI 0.92-0.97), P = 0.0002; triglycerides at 1.03 (95% CI 1.01-1.04), P < 0.0001; white blood cells at 1.74 (95% CI 1.26-2.40), P = 0.0008; and ferritin at 1.00 (95% CI 1.00-1.00), P = 0.0479 (Table 4).
| Odds of NAFLD | Odds of Fibrosis | |||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Demographics | ||||
| Age | 1.02 (1.00-1.05) | 0.0832 | 1.05 (0.99-1.11) | 0.1071 |
| Age ≥45 | 2.92 (0.85-10.05) | 0.0894 | 5.78 (0.69-48.21) | 0.1053 |
| Male | 2.75 (0.92-8.24) | 0.0713 | 4.65 (1.09-19.87) | 0.0379 |
| Hispanic | 1.67 (0.47-5.91) | 0.4289 | 3.26 (0.54-19.70) | 0.1971 |
| Anthropometric | ||||
| Height (cm) | 1.02 (0.95-1.09) | 0.6515 | 0.99 (0.90-1.08) | 0.7534 |
| Weight (kg) | 1.07 (1.04-1.11) | <0.0001 | 1.05 (1.02-1.08) | 0.0008 |
| BMI (kg/m2) | 1.26 (1.12-1.43) | 0.0002 | 1.21 (1.10-1.32) | <0.0001 |
| BMI >30 | 6.42 (2.40-17.16) | 0.0002 | 7.72 (1.65-27.39) | 0.0079 |
| SBP (mm Hg) | 1.03 (1.01-1.04) | 0.0108 | 1.04 (1.01-1.08) | 0.0172 |
| DBP (mm Hg) | 1.02 (0.99-1.05) | 0.1533 | 1.04 (1.00-1.08) | 0.0548 |
| Waist (cm) | 1.10 (1.05-1.15) | 0.0002 | 1.08 (1.03-1.13) | 0.0018 |
| Hip (cm) | 1.09 (1.04-1.14) | 0.0002 | 1.04 (1.00-1.08) | 0.0274 |
| Laboratory data | ||||
| Glucose (mg/dL) | 1.03 (1.01-1.05) | 0.0132 | 1.03 (0.99-1.07) | 0.1075 |
| Insulin (U/L) | 1.12 (1.00-1.26) | 0.0563 | 1.10 (0.97-1.24) | 0.1253 |
| HbA1c | 4.31 (1.75-10.65) | 0.0015 | 3.48 (1.04-11.61) | 0.0426 |
| HOMA-IR | 1.63 (1.08-2.44) | 0.0189 | 1.70 (1.04-2.79) | 0.0359 |
| AST (U/L) | 1.01 (0.98-1.04) | 0.6153 | 1.04 (1.00-1.09) | 0.0427 |
| ALT (U/L) | 1.03 (1.01-1.06) | 0.0104 | 1.03 (1.01-1.05) | 0.0046 |
| Alkaline phosphatase (U/L) | 1.00 (0.98-1.01) | 0.5761 | 1.01 (0.99-1.02) | 0.4051 |
| Albumin (g/dL) | 1.02 (0.37-2.85) | 0.9699 | 0.62 (0.09-4.25) | 0.6250 |
| GGT (U/L) | 1.01 (0.99-1.03) | 0.3642 | 1.03 (1.01-1.06) | 0.0175 |
| Total cholesterol (mg/dL) | 1.00 (0.99-1.01) | 0.5128 | 1.00 (0.99-1.02) | 0.6757 |
| HDL-cholesterol (mg/dL) | 0.95 (0.92-0.97) | 0.0002 | 0.90 (0.85-0.95) | 0.0002 |
| LDL-cholesterol (mg/dL) | 1.01 (1.00-1.02) | 0.1528 | 1.01 (0.99-1.03) | 0.3180 |
| Triglycerides (mg/dL) | 1.03 (1.01-1.04) | <0.0001 | 1.02 (1.01-1.03) | <0.0001 |
| WBC (×103/μL) | 1.74 (1.26-2.40) | 0.0008 | 1.27 (0.80-2.04) | 0.3122 |
| Hemoglobin (g/dL) | 1.45 (0.96-2.19) | 0.0788 | 1.01 (0.64-1.59) | 0.9581 |
| Hematocrit (%) | 1.13 (0.98-1.31) | 0.1025 | 1.01 (0.86-1.18) | 0.9357 |
| Platelets (×103/μL) | 1.00 (0.99-1.01) | 0.4749 | 1.00 (0.98-1.01) | 0.5051 |
| INR | 0.94 (0.35-2.55) | 0.9002 | 10.34 (2.03-52.69) | 0.0049 |
| Ferritin (ng/mL) | 1.00 (1.00-1.00) | 0.0479 | 1.00 (1.00-1.01) | 0.1294 |
- Odds ratios derived from generalized estimating equations (PROC GENMOD) in SAS software, version 9.4, to account for intrapair correlations within twinships.
- Significant P <0.10. A two-tailed P < 0.05 is considered statistically significant (in bold type).
- Abbreviations: AST, aspartate aminotransferase; GGT, gamma- glutamyl transpeptidase; INR, international normalized ratio; WBC, white blood cell count.
PREDICTORS OF FIBROSIS IN OVERALL COHORT
Using generalized estimating equations, significant odds ratios as predictors of fibrosis in our overall cohort included male gender at 4.65 (95% CI 1.09-19.87), P = 0.0379; weight at 1.05 (95% CI 1.02-1.08), P = 0.0008; BMI at 1.21 (95% CI 1.10-1.32), P < 0.0001; BMI >30 at 7.72 (95% CI 1.65-27.39), P = 0.0079; SBP at 1.04 (95% CI 1.01-1.08), P = 0.0172; DBP at 1.04 (95% CI 1.00-1.08), P = 0.0548; waist circumference at 1.08 (95% CI 1.03-1.13), P = 0.0018; hip circumference at 1.04 (95% CI 1.00-1.08), P = 0.0274; HbA1c at 3.48 (95% CI 1.04-11.61), P = 0.0426; HOMA-IR at 1.70 (95% CI 1.04-2.79), P = 0.0359; aspartate aminotransferase at 1.04 (95% CI 1.00-1.09), P = 0.0427; ALT at 1.03 (95% CI 1.01-1.05), P = 0.0046; gamma-glutamyl transpeptidase at 1.03 (95% CI 1.01-1.06), P = 0.0175; HDL cholesterol at 0.90 (95% CI 0.85-0.95), P = 0.0002; triglycerides at 1.02 (95% CI 1.01-1.03), P < 0.0001; and international normalized ratio at 10.34 (95% CI 2.03-52.69), P = 0.0049 (Table 4).
Discussion
MAIN FINDINGS
In this study, we used a well-characterized, prospective, community-dwelling twin cohort design to demonstrate that hepatic steatosis and hepatic fibrosis have statistically and clinically significant shared gene effects. This builds on our previous findings that hepatic steatosis and fibrosis are each individually heritable traits.9 We also demonstrated significant shared genetic effects between hepatic steatosis and fibrosis and a wide number of metabolic risk factors, including HDL, triglycerides, insulin resistance, and HbA1c. These results suggest a genetic basis underlying the pathogenesis of both hepatic steatosis and fibrosis and with metabolic risk factors. This is a paradigm-changing finding as most experts believe that hepatic steatosis is inconsequential and only hepatic fibrosis is associated with worse outcomes, including mortality and liver transplantation.5 The shared gene effects between hepatic steatosis and hepatic fibrosis suggest that development of hepatic steatosis may itself portend a worse outcome. However, the time horizon for hepatic steatosis to reach these adverse outcomes may be long, and studies with 10-20 years of follow-up may be needed to assess these outcomes. It also has implications in developing targeted therapies for the treatment of NASH. It provides a biological plausibility that reduction of hepatic steatosis over a sustained period of time may also influence the expression of genes associated with fibrosis progression/regression and may be viable target for the treatment of NASH.
IN THE CONTEXT OF THE PUBLISHED LITERATURE
Studies have shown that both hepatic steatosis9, 35, 36 and fibrosis9 are heritable traits. We build on the results of these studies to show additional heritabilities between hepatic steatosis and fibrosis. Additionally, NAFLD has been associated with metabolic risk factors,17-20 although the relative contributions of genetic versus environmental factors to these associations are unknown from these studies. While we have previously demonstrated genetic covariance between NAFLD and metabolic risk factors in a prospective twin study design, gamma-glutamyl transferase was used as a marker of hepatic steatosis and liver fat content was not measured directly.21 Additionally, no previous studies have demonstrated genetic covariance between hepatic fibrosis and metabolic risk factors. This is the first study to demonstrate genetic covariance between metabolic risk factors and both hepatic steatosis and fibrosis in a community-dwelling cohort of twins, with accurate quantification of steatosis and fibrosis throughout the liver achieved through the use of noninvasive MRI-based imaging techniques.
There are currently few effective medical therapies to manage NAFLD and its complications. Vitamin E and thiazolidinediones have been shown to improve hepatic steatosis in NAFLD patients.38-40 However, few treatments have been shown to be effective in reversing NAFLD-associated hepatic fibrosis. The genes PNPLA310, 41 and TM6SF242, 43 have been shown to modify the risks of hepatic steatosis and fibrosis, and other genetic pathways associated with steatosis, fibrosis, and metabolic traits remain to be elucidated. Future identification of these genetic pathways may lead to individualized, targeted therapies that may prevent and/or reverse hepatic steatosis and fibrosis.
STRENGTHS AND LIMITATIONS
The strength of this study lies in its use of a twin study design that allows for the evaluation of the heritability of steatosis, fibrosis, and metabolic risk factors. The cohort consisted of well-characterized, community-dwelling twins in which twins with conditions such as excessive alcohol use, use of steatogenic medications, viral hepatitis, and secondary causes of steatosis were systemically excluded. The use of MRI-PDFF allowed for detailed mapping and steatosis quantification throughout the liver, and the use of MRE allowed for an accurate, noninvasive way to quantify hepatic fibrosis.
However, this study is limited by the lack of biopsy, which remains the gold standard for diagnosing liver steatosis and fibrosis. While biopsies are limited by their interobserver variability and sampling bias, they allow for the diagnosis of lobular inflammation, hepatocyte ballooning, and NASH that cannot be diagnosed noninvasively. However, because it is unethical to perform liver biopsies in normal control patients with no suspicion of NAFLD, a study involving liver biopsies can only be performed if at least one twin has suspected NAFLD. Our use of noninvasive biomarkers instead of liver biopsy to assess hepatic steatosis and fibrosis allowed us to use a community-dwelling cohort of patients, rather than preselected patients with increased risk of NAFLD. Although MRI-PDFF has been shown to have high interreader reproducibility in nontwin studies44 and the interobserver variability of magnetic resonance readings in our study was minimized with only one analyst performing all the image analyses, the general interobserver variability of magnetic resonance readings in similar twin study designs remains unknown. Additionally, MRI-PDFF has been shown to be highly accurate for mapping hepatic steatosis throughout the liver without the sampling variability associated with liver biopsies, and MRE has also been shown to be highly accurate for the diagnosis of hepatic fibrosis, so we believe our noninvasive diagnostic tests can reliably measure steatosis and fibrosis.31-33
IMPLICATION FOR FUTURE STUDY
In this study, we demonstrate in a prospective, community-dwelling cohort of twins that patients with genetic susceptibility to hepatic steatosis also have genetic susceptibility to hepatic fibrosis. We also demonstrate that hepatic steatosis and fibrosis have shared genetic effects with metabolic risk factors. Additional studies with larger sample sizes will be needed to identify individual genes or pathways that may be implicated in hepatic steatogenesis and/or fibrogenesis. The identification of these genes may allow for further individualized, targeted therapy that may prevent and even reverse hepatic steatosis and fibrosis.
REFERENCES
Author names in bold designate shared co-first authorship.




