MS-based multi-dimensional metabolomics reveals protective effect of Polygalae Radix against metabolic disturbances in Alzheimer's disease mice
Dear Editor
The system-wide metabolic dysregulation is a central hallmark of Alzheimer's disease (AD).1-3 Growing evidence indicated that Polygalae Radix (PR) has ameliorative effects on memory deficits of AD.4, 5 However, the anti-AD effect of PR extract (PRE) via regulating metabolic disturbance is seldom investigated from the perspective of a system-wide level. In this study, matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI–MSI)-based spatial metabolomics and liquid chromatography-mass spectrometry (LC–MS)-based metabolomics were applied to explore the anti-AD effect of PRE via improving the system-wide metabolic disorders in APPswe/PSEN1dE9 (APP/PS1) double transgenic AD model mice.
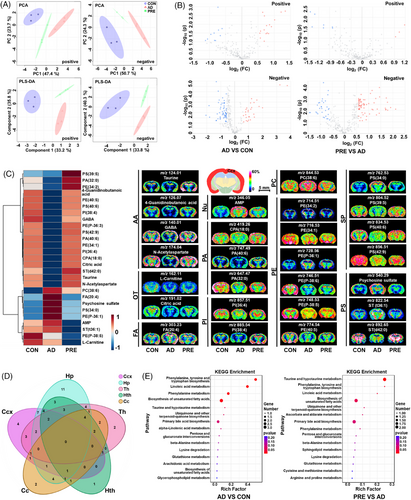
The chemical composition of PRE was first characterised by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC–MS/MS) (Figure S1 and Table S1). The results of behavioural and pharmacological experiments indicated that PRE treatment remarkably ameliorated the impairment of learning and memory in AD model mice, and significantly reduced the levels of amyloid-β (Aβ) plaques (Figure S2) and Aβ1–42 monomers, and increased the levels of acetylcholine and brain-derived neurotrophic factor in the cerebral cortex (Ccx) and hippocampus (Hp) of AD mice (Figure S3) (see the Supporting Information for details). Subsequently, MALDI–MSI was utilised to investigate the ameliorative effect of PRE against regional metabolic disturbances in AD mouse brains. Principal component analysis (PCA) and partial least squares discrimination analysis (PLS-DA) showed that the metabolic profiles in Ccx, Hp, corpus callosum (Cc), thalamus (Th) and hypothalamus (Hth) of PRE-treated mice were remarkably distinguished from AD, indicating that notable spatial metabolic disturbances could be changed by PRE (Figures 1A and S4). Tissue-specific differential metabolites between PRE-treated and AD model mice were identified in five brain regions. In Ccx, 59 differential metabolites were observed, and the levels of 27 out of 59 returned to control (CON) after PRE treatments (Figure 1B-1C and Table S2). The spatial distribution and level changes of representative metabolites in CCx before and after PRE treatment are shown in Figure 1C. Similarly, a total of 75, 59, 56 and 53 differential metabolites between the PRE-treated and AD model groups were identified in Hp, Th, Hth and Cc regions, respectively (Figure S5 and Tables S3–S6). Among them, the levels of 39 differential metabolites in Hp, 23 in Th, 23 in Hth and 19 in Cc had been reversed after PRE treatments (Figures S6 and S7). Therefore, MALDI–MSI results revealed that PRE treatment could regulate the drastic brain metabolic disturbances with a critical spatial metabolic heterogeneity (Figure 1D). According to KEGG reference pathways, PRE treatment regulated the imbalance of multiple metabolism pathways at different levels in five brain regions (Figures 1E, S8 and S9). A partial metabolic pathway network of selected significantly altered metabolites was mapped (Figure 2), demonstrating that PRE treatment corrected spatial metabolic disturbances in the AD model mouse brain in a region-specific manner.

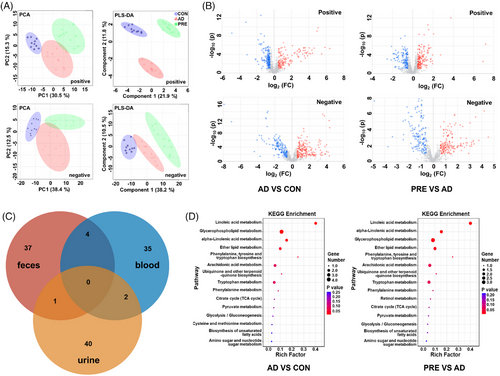
Moreover, blood, urine and faeces were analysed using UPLC–MS/MS to investigate the biofluid metabolic changes. PCA and PLS-DA results revealed three distinct groups of blood (Figure 3A). Similar results were observed in urine and faeces (Figure S10). OPLS-DA results showed that 41 differential metabolites in blood were identified between PRE-treated and AD groups, 43 in urine and 42 in faeces. PRE treatment reversed the abnormal levels of 28 differential metabolites in the blood, 33 in urine and 24 in faeces (Figures 3B and S11–S13 and Tables S7–S9). The similarities and differences of the reversed metabolites in blood, urine and faeces are shown in Figure 3C. KEGG pathway enrichment results showed that PRE treatment regulated the linoleic acid metabolism, alpha-linolenic acid metabolism, ether lipid metabolism to ameliorate metabolic disturbance in the blood of AD mice (Figure 3D). In urine, the disturbed pathways, including valine, leucine and isoleucine biosynthesis, aminoacyl-tRNA biosynthesis and phenylalanine metabolism, could be corrected by PRE treatments, and in faeces the disturbed pathways, including taurine and hypotaurine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, linoleic acid metabolism, could be corrected (Figure S14).
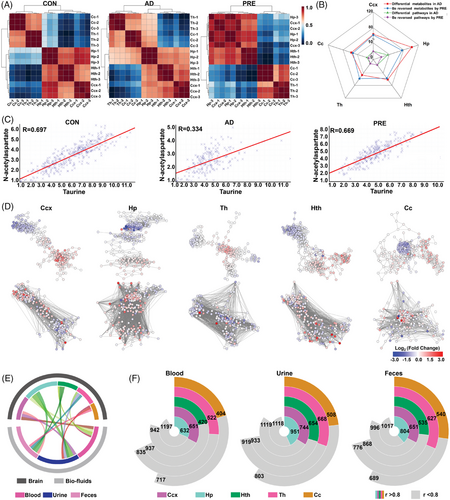
Finally, the Spearman-rank correlation matrix of five brain regions was plotted to investigate region-specific metabolic differences among three groups. In the CON group, high positive correlations were observed between Ccx and Hp (r > 0.8, p < .05) and moderate correlations between Cc and Th, Ccx and Hth, Hp and Hth (0.6 < r < 0.8, p < .05). However, the correlations among the Hp, Ccx, Th and Cc were totally altered in AD model mice. Notably, the high positive correlations between Ccx and Hp were recovered after PRE treatment and the moderate correlations between Hp and Cc, Hp and Th diminished (Figure 4A). Among five brain regions, Hp of AD model mice showed the most drastic change of regional metabolism in terms of the number of altered metabolites and metabolic pathways, followed by the Ccx (Figure 4B). The metabolite–metabolite correlation matrices among the three groups showed that AD model mice exhibited a notable reduction in both high positive and negative correlations among metabolites. Furthermore, there was a significant alteration in correlation patterns in PRE-treated mice (Figure S15). For example, the level of taurine was positively correlated with that of N-acetylaspartate in the Ccx and Hp of CON mice (R = 0.697 and 0.657, respectively), but the correlation was severely diminished in the AD model group (R = 0.334 and 0.380, respectively), whereas the correlation was significantly recovered in PRE-treated group (R = 0.669 and 0.625, respectively) (Figures 4C and S16). Furthermore, metabolic networks were constructed for each brain region, with nodes representing metabolites and edges representing correlations (R > 0.90) (Figure 4D). The results were consistent with metabolite–metabolite correlation matrices in which the altered correlations in AD model mice were significantly changed by PRE treatment (Figures 4D and S15). To evaluate the system-wide metabolic homeostasis, the correlation between the reversed metabolite levels in brain and biofluids (blood, urine and faeces) of PRE-treated mice was analysed. As shown in Figure 4E,F, 657 significant metabolite–metabolite correlations between blood and Ccx were observed, 951 between urine and Hp and 804 between faeces and Hp (r > 0.80, p < .05) (see the Supporting Information for details). These results indicated that Hp and Ccx regions might be the most susceptible to AD, and PRE treatment had a beneficial effect by correcting the metabolic disturbance that occurred in Hp and Ccx.
Overall, the regulatory effect of PRE on the disturbances of system-wide metabolic homeostasis in AD mice was demonstrated by MS-based multi-dimensional metabolomics. Spatial metabolomics revealed that PRE treatment significantly reversed the brain region-specific metabolic disturbances in AD mice and improve the metabolic coordination among five brain regions. Meanwhile, the perturbed metabolic profiles in the blood, urine and faeces of AD mice were significantly improved after PRE treatment, particularly concentrated in pathways associated with glycerophospholipid and sphingolipid metabolism. More importantly, PRE significantly improved the metabolic coordination between brain regions and blood, urine and faeces in AD mice, especially the inter-metabolite correlation between blood, urine, faeces and the Hp and Ccx regions, indicating that PRE could alleviate disturbance of system-wide metabolic homeostasis via regulating the metabolism coordination between central and peripheral system. Our findings may provide a new strategy for the multi-targeted treatment of AD.
AUTHOR CONTRIBUTIONS
Conceptualisation, investigation, methodology, validation and writing—original draft: Yanwen Chen. Investigation, methodology and validation: Lisha Zhao. Methodology and validation: Shuo Cai. Investigation, methodology and validation: Yuchen Zou. Conceptualisation, investigation, methodology and validation: Weiwei Tang. Conceptualisation, supervision, writing—review and editing: Bin Li. All authors reviewed the manuscript and approved the submitted version.
ACKNOWLEDGEMENTS
The authors have nothing to report.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China (No. 82374028, No. 82304894, and No. 81773873) and the Jiangsu Funding Program for Excellent Postdoctoral Talent. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
ETHICS STATEMENT
All animal protocols were approved by the Institutional Animal Care and Use Committee of China Pharmaceutical University, which are in accordance with the National Institutes of Health guidelines.




