CRBN is downregulated in lung cancer and negatively regulates TLR2, 4 and 7 stimulation in lung cancer cells
Mi-Jeong Kim, Ji Su Lee, Ji Young Kim and Bongkum Choi contribute equally to this work.
Dear Editor,
Cereblon (CRBN) has been identified as a primary target of immunomodulatory drugs in multiple myeloma.1 Herein, for the first time, we demonstrate that CRBN expression is functionally involved in lung cancer progression through the regulation of autophagy by toll-like receptor (TLR)2, TLR4 and TLR7. TLR signalling is associated with the induction of autophagy and plays a pivotal role in the progression and pathogenesis of lung cancer.2 Our study suggests that CRBN can be a potent prognostic marker for lung cancer and provides important implications in clinical and translational lung cancer biology.
To get insight into the association of CRBN with lung cancer, we utilized data from the TCGA (The Cancer Genome Atlas) (GEPIA, http://gepia.cancer-pku.cn/detail.php?gene = CRBN) and a cohort of NSCLC patients (n = 18). GEPIA database revealed that CRBN was downregulated in lung adenocarcinoma and lung squamous cell carcinoma (Figures S1 and 1A, LUSC, *p < .05). Moreover, CRBN was downregulated in lung tumour tissues (LTTs) of 18 NSCLC patients compared to matched lung normal tissues (LNTs) (Figure 1B and, Table S1). To see whether CRBN expression is associated with a defined set of genes related to cancer in LTTs of NSCLC patients, we selected the top seven LTTs (<−1.5-fold decrease of CRBN) (Figure 1B, red bars) and performed gene set enrichment analysis (GSEA). The 15 enrichment gene set plot related to oncogenic signature and cancer was enriched in the top 7 LTTs as compared to matched LNTs of NSCLC patients (Figure S2). To decipher the gene association between CRBN expression and lung cancer progression in the 7 LTTs, we aligned 500 downregulated or upregulated genes based on the data of LTT36 with the most downregulation (Figure S3A and Table S2, downregulated genes; Figure S4A and Table S3, upregulated genes). We further sorted downregulated or upregulated genes in all seven LTTs (Figures S3B–D and S4B,C). Then, we accessed PubMed to confirm whether these genes have been reported in lung cancer proliferation, migration or invasion and progression. Among downregulated genes (Figure S3B–D), 17, 11 and 21 genes have been involved in anti-lung cancer proliferation (Figure 1C, Table S4), anti-lung cancer migration or invasion (Figure 1D, Table S5) and anti-lung cancer progression (Figure 1E, Table S6), respectively. Among upregulated genes (Figures 1F and S4B,C), 13, 10 and 4 genes have been involved in lung cancer proliferation (Figure 1G, Table S7), migration or invasion (Figure 1H, Table S8) and progression (Figure 1I, Table S9), respectively. We observed a similar pattern in the other 11 LTTs (Figures S5A–C and S5D–F). To examine the role of CRBN in lung cancer cells, we generated CRBN-knockout (CRBNKO) A549 (Figure 1J) and H1299 lung cancer cells (Figure S6) and performed in vitro and vivo cancer progression assays. CRBNKO A549 cells significantly enhanced cancer migration (Figure 1K), invasion (Figure 1L), single-cell migration (Figure 1 M), anchorage-dependent (Figure 1N) and anchorage-independent colony formation (Figure 1O). We also observed a similar pattern in CRBNKO H1299 cells (Figure S7A–D). Notably, as an in vivo model, NOD/SCID/IL2rγnull (NSG) mice engrafted with CRBNKO A549 cells markedly increased the tumour sizes and masses (Figure 1P,Q, CRBNKO A549 vs. Ctrl A549). H&E staining data revealed that the number of neoplastic epithelial cells was higher in tumour and metastasized LTTs derived from mice injected with CRBNKO A549 (Figure S8A,B, tumour; Figure S8C,D, metastasized lung tumour) compared to those of Ctrl A549, demonstrating that (i) CRBN downregulation is associated with lung cancer; (ii) CRBN-deficiency promotes in vitro and vivo lung cancer progression.
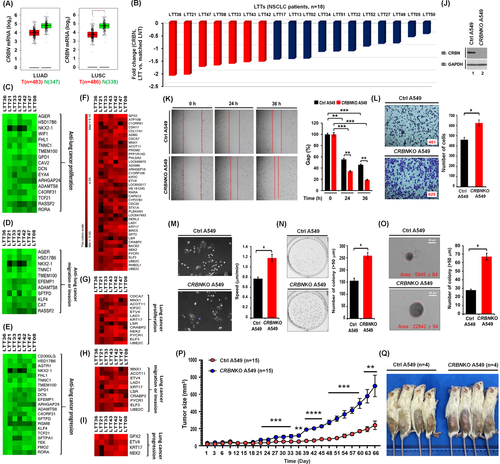
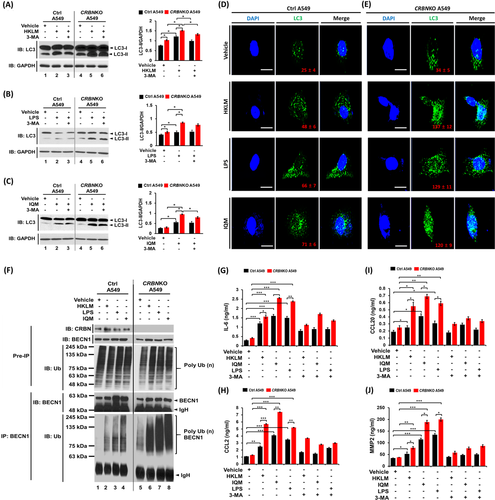
Growing evidence suggests that intrinsic and extrinsic factors, including spontaneous mutations of genes and TLRs, play pivotal roles in lung cancer development and progression.2-7 Importantly, TLR2, TLR4 and TLR7 are expressed in lung cancer and associated with lung cancer progression.5-7 Additionally, TLR4- and TLR3- induced autophagy facilitates migration and invasion of lung cancer cells.2 We previously reported that CRBN negatively regulates TLR4 signalling through the attenuation of ubiquitination of TRAF6, and autophagy activation by inhibiting the ubiquitination of BECN1.8, 9 Therefore, we sought to determine whether CRBN is involved in lung cancer progression by TLR2, TLR4 and TLR7 signals associated with autophagy. We first examined if TLR2, TLR4 and TLR7 signals are associated with genes related to lung cancer progression and autophagy. We treated A549 and H1299 cells with vehicle as a control, TLR2 (heat-killed Listeria monocytogenes, HKLM), TLR4 (lipopolysaccharide, LPS) or TLR7 (imiquimod, IQM) agonists and performed RNA-sequencing analysis. The GSEA of transcriptional profiles revealed that 15 enrichment gene set plots related to cancer gene neighbourhoods and cancer modules, ontology and oncogenic signature were enriched in A549 and H1299 treated with TLR4 agonist compared to those with the vehicle (Figure S9A–C, A549; Figure S9D–F, H1299). To examine the association of genes related to lung cancer progression and autophagy in response to TLR stimulation, we arranged 500 upregulated genes (Figure S10A and Table S10 in A549; Figure S10B, Table S11 in H1299) or 500 downregulated genes (Figure S10C and Table S12 in A549; Figure S10D and Table S13 in H1299) based on the data of TLR4 stimulation. Then, we further sorted upregulated (Figure S11A and Table S14 in A549; Figure S11B and Table S15 in H1299) or downregulated (Figure S11C and Table S16 in A549; Figure S11D and Table S17 in H1299) genes in all three TLR stimulations. Among upregulated genes, 12 genes (Figure 2A and Table S18) in A549 cells and 21 genes (Figure 2B and Table S19) in H1299 cells have been involved in lung cancer progression. In the list of downregulated genes, 10 genes (Figure 2C and Table S20) in A549 cells and 11 genes (Figure 2D and Table S21) in H1299 cells have been involved in anti-lung cancer progression. Notably, 13 genes upregulated in A549 or H1299 cells have been functionally associated with autophagy (Figure 2E, Table S22). Moreover, enrichment gene sets regulating autophagy, such as BECN1 (Figure S12A), MTOR (Figure S12B) and AKT-MTOR (Figure S12C) were enriched in A549 treated with TLR4 agonist compared to those with the vehicle. These results suggest that TLR2, TLR4 and TLR7 stimulation increase gene signatures related to lung cancer progression and autophagy (Figure 2F).

We next examined whether CRBN is functionally involved in lung cancer progression through the regulation of autophagy by TLR stimulation. Under the TLR2 (HKLM), TLR4 (LPS) or TLR7 (IQM) stimulation, cell migration and invasion were significantly enhanced in CRBNKO A549 cells (Figure 2G,H, HKLM; Figure 2I,J, LPS; Figure 2K,L, IQM: CRBNKO A549 vs. Ctrl A549). Marked inhibition was observed in the presence of autophagy inhibitors, 3-MA or CQ (Figure 2G–L, HKLM, LPS or IQM agonist + 3-MA or CQ vs. TLR agonist alone). Importantly, LC3-II levels and LC3 puncta that represent induction of autophagy were significantly enhanced in CRBNKO A549 treated with TLR agonists (Figure 3A–C, LC3-II levels; Figure 3D,E, LC3 puncta). TLR4 signalling induces autophagy through the BECN1 ubiquitination by TRAF6.2, 8-10 Then, we examined whether CRBN-deficiency directly affects the BECN1 ubiquitination by TLR2, TLR4 and TLR7 stimulation. BECN1 ubiquitination was markedly elevated in CRBNKO A549 cells treated with HKLM, LPS or IQM as compared to those of Ctrl A549 cells (Figure 3F, lane 6 vs. lane 2 in HKLM; lane 7 vs. lane 3 in LPS; lane 8 vs. lane 4 in IQM). IL-6, CCL2, CCL20 and MMP2 productions are necessary for enhanced migration and invasion of lung cancer cells upon TLR activation.2 We found that CRBNKO A549 cells markedly increased IL-6, CCL2, CCL20 and MMP2 in response to three TLR agonists compared to those of Ctrl A549 (Figure 3G, IL-6; Figure 3H, CCL2; Figure 3I, CCL20; Figure 3J, MMP2: CRBNKO A549 vs. Ctrl A549), whereas marked inhibitions were observed in the co-treatment of autophagy inhibitors (Figure 3G–J, 3-MA). Lastly, we found that CRBNKO A549 cells treated with TLR agonists significantly increased single-cell mobility and the number of colonies compared to those of Ctrl A549 cells (Figure 4A,B, single-cell mobility; Figure 4C,D, number of colonies).
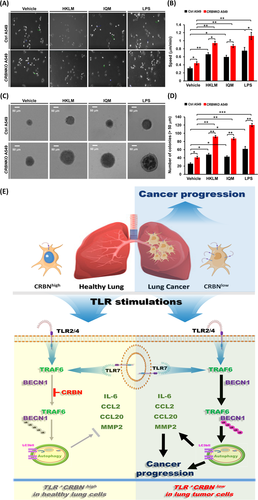
In summary, CRBN is downregulated in lung cancer cells and associated with lung cancer progression (Figure 4E, upper right). Our study demonstrates the association between TLR stimulation and gene signatures related to lung cancer progression and autophagy. CRBN inhibits the BECN1 ubiquitination to induce autophagy and attenuates the production of IL-6, CCL2, CCL20 and MMP2 cytokines in response to TLR stimulations in healthy lung cells expressing CRBN (CRBNhigh, Figure 4E, down left). In lung cancer cells with downregulated CRBN (CRBNlow, Figure 4E, down right), engagements of TLRs enhance autophagy induction through the increases of BECN1 ubiquitination and the production of IL-6, CCL2, CCL20 and MMP2 cytokines, eventually facilitating lung cancer progression. Taken together, our clinically comparative results and functional investigations of CRBN in lung cancer progression will potentially contribute to translational approaches for lung cancer intervention.
ACKNOWLEDGEMENT
We would like to thank Hyehwa Forum members for their helpful discussion. This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (NRF-2021R1F1A1049324 and NRF-2021R1A2C1094478), Ministry of Science ICT and Future Planning (MSIP) funded by the Korean Government (NRF-2016R1A5A2945889).
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
CONSENT FOR PUBLICATION
All authors agree to publish this article.




