Expression of CD49f defines subsets of human regulatory T cells with divergent transcriptional landscape and function that correlate with ulcerative colitis disease activity
Lead contact.
Abstract
Objective
Adoptive regulatory T cell (Treg) therapy is being trialled for the treatment of different autoimmune disorders, including inflammatory bowel diseases (IBD). In-depth understanding of the biological variability of Treg in the human blood may be required to improve IBD immune monitoring and treatment strategies.
Methods
Through a combination of quantitative proteomic, multiparametric flow cytometry, RNA-sequencing data analysis and functional assays on Treg enriched from the blood of ulcerative colitis (UC) patients and healthy controls, we investigated the association between CD49f expression, Treg phenotype and function, and UC disease activity.
Results
High-dimensional analysis and filtering defined two distinct subsets of human Treg based on the presence or absence of CD49f with divergent transcriptional landscape and functional activities. CD49f negative (CD49f−) Treg are enriched for functional Treg markers and present significantly increased suppressive capacity. In contrast, CD49fhigh Treg display a pro-inflammatory Th17-like phenotype and accumulate in the blood of patients with UC. Dysregulation on CD49f Treg subsets in patients with UC correlate with disease activity.
Conclusion
Overall, our findings uncover the importance of CD49f expression on Treg in physiological immunity and in pathological autoimmunity.
Introduction
Immune suppression through regulatory T cells (Treg) is pivotal for maintaining body homeostasis, controlling exaggerated immune responses against pathogens, and the prevention of immune cells attacking healthy tissue in the cases of autoimmunity, allergy, allograft rejection and foetal rejection during pregnancy.1 While the overall Treg cell population is defined as CD4+ T cells bearing a CD25+FoxP3+CD127– phenotype, Treg found in peripheral circulation are highly heterogenic and have diverse function. At least 22 phenotypically different Treg subsets have been identified by mass cytometry in humans.2 Furthermore, many activated conventional CD4+ T cells (conv CD4+) can also express CD25 and FoxP3 at low levels, which cloud the specific identification of Treg.3 It is possible that a comprehensive multi-‘omic’ approach associating both proteomic and transcriptome analysis could lead to more precise characterisation of the various Treg subsets providing new insights into Treg mechanisms that guide homeostasis in health and dysfunction in disease.
Because of their multiple suppressive mechanisms, Treg represent a promising strategy for adoptive cell therapy for chronic inflammatory diseases. Treg are critical for commensal tolerance in the intestine, and a lack of intestinal tolerance can lead to chronic inflammation including inflammatory bowel diseases (IBD) consisting mainly of Crohn’s disease (CD) and ulcerative colitis (UC).4-6 Evidence from both animal models and patients supports the idea that Treg therapy would be beneficial in the context of IBD. Treg have been expanded from patient’s blood and safely used in recent phase 1 studies designed for the treatment of CD, type 1 diabetes mellitus, lupus and autoimmune hepatitis.7-9 Despite this great promise, the therapeutic use of Treg has been hampered by the biological variability of Treg populations in the peripheral blood. Effector Treg are heterogeneous and unstable following expansion; however, they do demonstrate increased suppressive function, higher efficacy and specificity in controlling immune responses compared with resting Treg.10
Besides the loss of Treg suppressive function, infiltration of pro-inflammatory T-helper 17 (Th17) cells is also implicated in the pathogenesis of IBD.11 Interestingly, Treg differentiation is tightly linked to the development of Th17 cells, an effector T cell subset involved in the induction of inflammation and implicated in autoimmune tissue injury through the production of interleukin-17A (IL-17A) and other pro-inflammatory cytokines.12 Whereas both the induction of peripheral Treg from resting CD4+ T cells and the maintenance and function of thymus-derived natural Treg are dependent on transforming growth factor beta (TGF-β) signalling, the presence of IL-6 inhibits TGF-β-mediated FoxP3 induction and drives cells towards a Th17 phenotype. A subset of Treg cells expressing the Th17-associated markers’ retinoid-related orphan receptor-gamma t (RORγt) and chemokine receptor 6 (CCR6), in addition to FoxP3, have also been reported in vivo and is increased in the intestinal mucosa and among peripheral blood mononuclear cells (PBMC) circulating in patients with IBD in relation to healthy controls.12-14 However, the mechanisms that underpin the development of these Th17-like Treg cells are still under debate because of the high Treg cell plasticity, which can be detrimental in the setting of autoimmune diseases.
Several T cell subsets express adhesion receptors known as integrins, such as CD49a, CD49b, CD49d and CD49f, which have been reported to modulate various aspects of T cell biology including cell differentiation, migration and functionality.15-18 It is possible that CD49f (integrin alpha 6) expression on CD4+ T cells is associated with the pathogenesis of IBD, as CD49f is increased on the surface of circulating conv CD4+ cells that migrate out of the colonic mucosa of patients with active IBD.19
In order to assess the impact of CD49f expression on Treg-mediated immune responses in health and disease, we investigated the association between CD49f expression, Treg phenotype and function, and clinical outcomes in patients with IBD. Comparative proteomics between Treg and conv CD4+ cells reveal CD49f to be divergently expressed among circulating Treg. Using high-dimensional analysis and filtering, we define two subsets of CD4+CD25high Treg, which have been shown to exhibit a strong regulatory function in humans,20 based on the presence or absence of CD49f, with divergent transcriptional landscape and functional activities. Our data reveal that CD49f negative (CD49f−) Treg exert high suppression on conv CD4+ cell proliferation, associated with elevated expression of FoxP3 and the immune checkpoint receptors, CD39 and CTLA4. In contrast, CD49fhigh Treg produce abnormal levels of IL-17A under TCR-mediated activation, concurrently expressing higher levels of CCR6, and are increased in PBMC of patients with UC compared with healthy controls. Notably, an elevated CD49fhigh/CD49f− effector Treg ratio (CD49feR) in patients’ blood is a predictor of active disease in UC. Taken together, our findings demonstrate that CD49f expression on Treg impacts human physiological immunity and influence the development of IBD and possibly other autoimmune disorders.
Results
CD49f is divergently expressed among human regulatory T cells
Treg cells are generally identified as a CD4+ T cell subset with suppressive properties presenting high phenotypic and functional diversity.2 To allow better characterisation of Treg in humans, we set out to identify differentially expressed surface proteins between Treg (CD4+CD25high) and conv CD4+ cells (CD4+CD25−) using comparative proteomics. Treg with high purity were obtained from human PBMC through sequential magnetic and flow cytometry cell sorting (FACS) (Figure 1a). As expected, all CD4+CD25high cells were CD127−FoxP3high, whereas FoxP3 expression on conv CD4+ cells was similar to unstained controls21 (Figure 1a). We conducted label-free quantitative proteomics using data-dependent acquisition (DDA-MS). Inspection of the maxLFQ normalised intensity values showed missing values in < 10% of proteins in each sample (Supplementary figure 1a), confirming the acquisition of high-quality DDA-MS data for unambiguous label-free quantification. In total, we identified 4,177 protein groups at 1% of false discovery rate (FDR) (Supplementary figure 1b). 2,788 proteins were quantified using single UniProt accessions with at least 2 unique and razor peptides in more than 50% of the samples and thus selected for differential expression (DE) analysis (Supplementary figure 1b). Most of these proteins were quantified based on intensities of more than 5 peptides (Supplementary figure 1c) and showed distribution pattern common to all the samples analysed (Supplementary figure 1d). Hierarchical cluster analysis based on Euclidian distance clearly separated the CD4+ T cell proteomic data in two groups according to the subset analysed (Supplementary figure 1e). This was further confirmed by principal component analysis (PCA), in which two clear clusters were observed in the first principal component (Figure 1b). Most of the proteins within each subset had < 2% co-variability (Supplementary figure 1f), verifying the consistency and reproducibility of the obtained label-free quantitative DDA-MS data. In addition, we used Ingenuity Pathway Analysis (IPA, Qiagen bioinformatics, USA) to characterise the subcellular distribution of the proteins identified. As expected, most of the proteins detected from whole-cell lysates were derived from the cytoplasm and nucleus, whereas 180 proteins (˜6% of the total proteins quantified) were annotated as plasma membrane proteins and considered of interest as potential uncharacterised Treg surface markers (Figure 1c and Supplementary table 1). Statistical analysis identified 227 proteins as DE (FDR < 0.05 and log2FC > 1 or < −1) between donor-matched Treg and conv CD4+ cells and indicated that only 10% of the global Treg proteome was significantly different from conv CD4+ (Supplementary table 2). Of the DE proteins, 72% (n = 166) were upregulated in Treg cells, including FoxP3 with a log2-fold change of 6.29 (Figure 1d, Supplementary table 2). As a candidate Treg surface marker, we selected the plasma membrane protein CD49f, which showed a 3.12 log2-fold increase in relation to conv CD4+ (Figure 1d). Subsequent flow cytometric validation using a anti-CD49f monoclonal antibody revealed that CD49f is heterogeneously expressed in human Treg, allowing the identification of 3 distinct Treg populations characterised as CD49f−, CD49fdim and CD49fhigh (Figure 1e). In accordance with the proteomic data, both CD49f mean fluorescence intensity (MFI) and the fraction of CD49fhigh cells were significantly increased in Treg in comparison with conv CD4+ cells (Figure 1f).
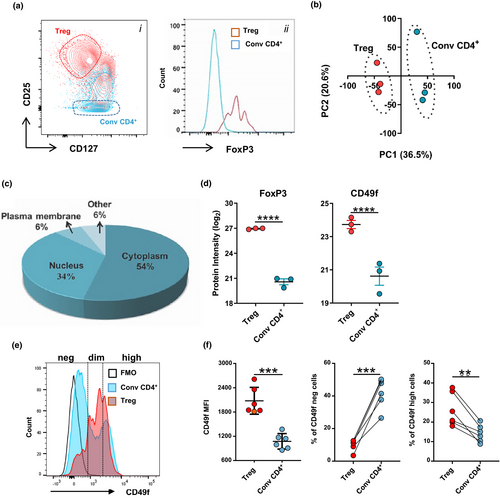
Thus, CD49f might define a unique subset of Treg with unexplored functions.
CD49f impacts Treg immunosuppressive ability and IL-17A production
We next wanted to understand the effect of CD49f expression on Treg function. For this, we sequentially sorted CD4+CD25high Treg from PBMC of five healthy donors (Supplementary figure 2a) and further stratified the cells based on CD49fhigh and CD49f− expression (Figure 2a). The immunosuppressive ability of CD49fhigh/− CD4+CD25high Treg was measured in vitro using a suppressive assay of autologous conv CD4+ cell proliferation in the presence of OKT3 antibodies (1 μg mL−1) and irradiated allogenic PBMC. Stimulated conv CD4+ cells without Treg were cultured in the same assay for definition of appropriate controls (Figure 2b). An increased proliferation of conv CD4+ cells was observed when the cells were co-cultured in the presence of CD49fhigh Treg in comparison with CD49f− cells. This effect was detected across multiple Treg: conv CD4+ cell ratios analysed (Figure 2b and c). CD49fhigh Treg showed a suppressive potential similar to total Treg, which was detected only when cells were cultured in a Treg: conv CD4+ cell ratio below 1:8 (Figure 2c, Supplementary figure 2b). Contrarily, CD49f− Treg were highly suppressive even when cells were cultured in a Treg: conv CD4+ cell ratio of 1:16 (Figure 2c). CD49f− Treg from all donors consistently presented increased ability to suppress CD4+ T cell proliferation, averaging 65.8 ± 6.89% versus 49.4 ± 3.37% of suppression observed in the CD49fhigh fraction (Figure 2d).
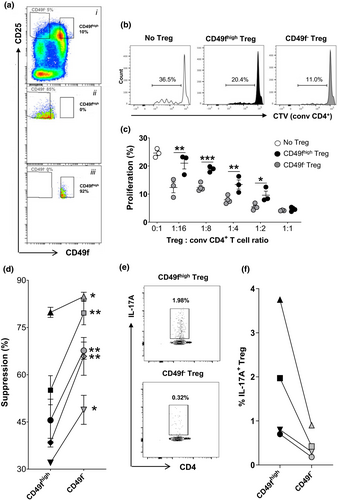
Based on previous studies indicating the existence of Treg that have the capacity to produce pro-inflammatory cytokines while retaining FoxP3 expression,12, 14 we sought to investigate an association between interleukin-17A (IL-17A) and interferon gamma (IFNγ) production by Treg and CD49f expression. Enriched CD49fhigh and CD49f− Treg were activated with CD3/CD28 Dynabeads in the presence of human recombinant IL-2 and analysed by flow cytometry for intracellular expression of IL-17A and IFNγ. In all donors evaluated, the proportion of IL-17A+ cells was 3- to 5-fold higher in CD49fhigh versus CD49f− Treg, comprising 1.8 ± 0.7% and 0.45 ± 0.16% of the cells analysed cells, respectively (Figure 2e and f). Similar to CD49f− Treg, only 0.6% of total Treg expressed IL-17A in the same experiment (Supplementary figure 2c). No association between CD49f and IFNγ expression was observed in activated Treg (Supplementary figure 2d), and most of IL-17A+ cells did not co-express IFNγ (Supplementary figure 2e). Of note, only a residual fraction of CD49fhigh Treg expressed IFNγ under activation in comparison with conv CD4+ cells (data not shown), evidencing that rather than representing a population of activated CD4+ T cells that contaminates Tregs, CD4+CD25high cells expressing high levels of CD49f represent a distinct population of IL-17A-producing Treg bearing an effector phenotype.
Taken together, our data show that CD49f expression impacts Treg immunosuppressive abilities and IL-17A production.
RNA-sequencing uncovers distinct subsets of regulatory T cells defined by CD49f expression
To comprehensively profile the relevant immune pathways associated with CD49fhigh and CD49f− Treg, we performed next generation RNA sequencing (RNA-Seq) on the two populations using high purity flow cytometry sorting of Treg from the peripheral blood of healthy individuals. We compared the transcriptional profiles between the sorted subsets, revealing two distinct Treg populations by PCA, in which two clear clusters were observed in the first principal component (Figure 3a). Transcriptional differences between CD49f− and CD49fhigh cells were further confirmed by hierarchical cluster analysis (Figure 3b). From the 17,936 genes identified, only 3.72% (n = 668) were DE between donor-matched CD49fhigh and CD49f− cells (FDR < 0.05, absolute log2FC > 1) (Supplementary figure 3a, Supplementary table 3) and 225 genes were upregulated in CD49fhigh Treg in comparison with their negative controls (Supplementary figure 3b). Gene expression differences were found in key molecules relevant for Treg effector function. CD49fhigh Treg expressed higher levels of genes associated with Th17 effector cytokine signalling, including RORC, CCR6, RORA, GPR65 and MYC in comparison with CD49f− controls. In contrast, CD49f− Treg presented increased expression of classical Treg markers including CTLA4, ENTPD1 (CD39), ICOS, LAG3 and FOXP3 (Figure 3c). Consistently, gene set enrichment analysis (GSEA) of a gene list derived from comparing Treg and CD4+ Th17 cells in healthy human PBMC (GSE107011) revealed that CD49fhigh Treg gene expression profile corresponded to the Th17 signature, while genes upregulated in CD49f− Treg overlapped with classical Treg gene signature (Figure 3d). No enrichment of Th1 signature was observed in the transcriptome of CD49fhigh Tregs (Supplementary figure 3c).
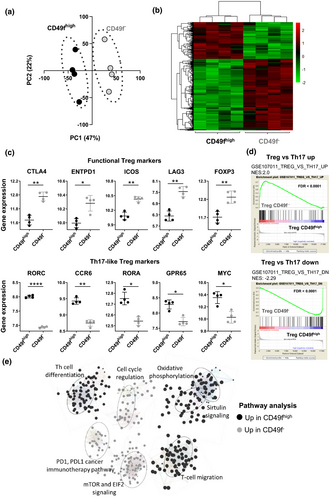
To identify altered relationships and pathways activity in these Treg subsets, DE genes were evaluated using IPA core analysis. Pathways upregulated in CD49fhigh Treg were those involved in the oxidative metabolism, cell migration, sirtuin signalling and T-helper cell differentiation. In comparison, CD49f− Treg were enriched for genes associated with cell cycle regulation, immune checkpoint modulation and mTOR and EIF2 signalling, which are critical regulators of Treg homeostasis and function (Figure 3e).
These data further validate the functional divergence between CD49fhigh and CD49f− Treg biology via significant transcriptional differences.
CD49f is associated with divergent effector regulatory phenotype and function in Treg
Next, we investigated the impact of CD49f expression on distinct subsets of Treg. As previously described, the simultaneous assessment of FoxP3 and CD45RA allows for the identification of three different subpopulations of human FoxP3-expressing CD4+ T cells: resting Treg (CD45RA+FoxP3low), effector Treg (CD45RA−FoxP3high) and FoxP3+ non-Treg cells (CD45RA−FoxP3low), which produce pro-inflammatory cytokines and lack suppressive capacity.21, 22 In this aim, we sought to validate DE markers originally identified by RNA-Seq in the subsets of CD49f− and CD49fhigh Treg using flow cytometric analysis. As expected, the degree of FoxP3 expression in all subpopulations analysed was proportional to CD25 expression21 (Figure 4a). CD4+ T cells expressing the highest levels of CD25 were also CD45RA−FoxP3high and therefore classified as effector Treg, whereas FoxP3+ non-Treg and resting Treg were part of the CD25 intermediate population (Figure 4a). CD4 MFI was similar among the three subsets analysed, but the CD25, FoxP3, CD39, CTLA4 and CCR6 MFI were increased in effector cells in relation to both resting Treg and FoxP3+ non-Treg (Supplementary figure 4a). Whereas CD49f fluorescence intensity did not differ between effector and resting cells (Figure 4b), CD49f specifically impacted the phenotype of effector Treg. In accordance with the RNA-Seq data, we observed an increased CD39, CTLA4 and FoxP3 MFI in CD49f− versus CD49fhigh effector cells (Figure 4c and d). In contrast, CCR6 MFI directly correlated with the level of CD49f expression on effector Treg (Figure 4c and d, Supplementary figure 4b). CD49f expression was also associated with CTLA4, CD39 and CCR6 MFI in FoxP3+ non-Treg (Figure 4c). CD49fdim cells expressed intermediate levels of each marker quantitated in both effector and FoxP3+ non-Treg cells. Resting Treg expressed similar levels of CD39, CTLA4, FoxP3 and CCR6 to FoxP3+ non-Treg (Supplementary figure 4b), which did not correlate with CD49f expression (Figure 4c). CD49f did not correlate with CD25 and CD127 MFI in either effector or resting Treg (Supplementary figure 4b).
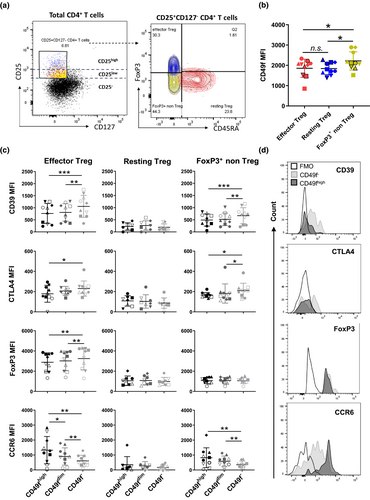
Thus, combined validation using different platforms indicates that CD49f impacts Treg effector function and is a lead target for Treg investigation.
CD49f expression on effector regulatory T cells correlates with disease activity in patients with ulcerative colitis
Because CD49f expression on effector Treg could potentially impact autoimmune diseases in which Treg play a role, and CD49f has been reported to modulate CD4+ T cell homing during IBD,19 we hypothesised that CD49f expression on circulating human Treg may be altered in autoimmune conditions such as IBD. To evaluate this, we characterised CD49f expression using flow cytometry in circulating Treg from a cohort of patients with UC who presented active or non-active disease at time of sampling (Table 1) and age-matched volunteer healthy controls. We noticed a trend towards reduction of total Treg in patients with UC (Figure 5a). While conv CD4+ cells were reduced in patients with active disease in comparison with healthy controls, the total fraction of Treg did not associate with UC disease activity (Supplementary figure 5a and b). Interestingly, CD49fhigh cells were significantly enriched in the effector Treg subset from patients’ blood in comparison with healthy controls (Figure 5b). While CD49fhigh cells accumulated in patients with active versus non-active disease, a decrease in CD49f− effector Treg was detected in patients with UC presenting active disease in relation to healthy controls (Figure 5c and d). Notably, the ratio of CD49fhigh/CD49f− effector Treg (CD49feR) in the peripheral blood significantly correlated with UC disease activity (R = 0.275; P = 0.004) (Figure 5e and f). A minor association between CD49f expression and UC disease activity was observed in resting Treg (Supplementary figure 5c–e), while no association was observed in FoxP3+ non-Treg (Supplementary figure 5e).
| Disease severity at time of sampling | ||||
|---|---|---|---|---|
| None (n = 12) | Mild (n = 5) | Moderate (n = 6) | ||
| Gender | ||||
| Male | 3 (25%) | 5 (100%) | 3 (50%) | |
| Female | 9 (75%) | 0 | 3 (50%) | |
| Age at diagnosis (years) | ||||
| Median | 25 | 27 | 28 | |
| Range | 15–56 | 12–33 | 17–39 | |
| Disease duration (years) | ||||
| Median | 9.6 | 8.6 | 6.2 | |
| Range | 0–44 | 1–19 | 2–17 | |
| Treatment | ||||
| 5-ASA | 1 (8.5%) | 3 (60%) | 4 (66%) | |
| Immunomodulators (IM) | 1 (17%) | |||
| Corticosteroid (CS) | 1 (8.5%) | |||
| 5-ASA + IM | 4 (33.5%) | 1 (20%) | ||
| 5-ASA + CS | 1 (17%) | |||
| Vedolizumab | 1 (8.5%) | 1 (20%) | ||
| None | 5 (41%) | |||
- 5-ASA, 5-aminosalicytes; CS, corticosteroid; IM, immunomodulators (Thioguanine 6TG, Mercaptopurine, azathioprine).
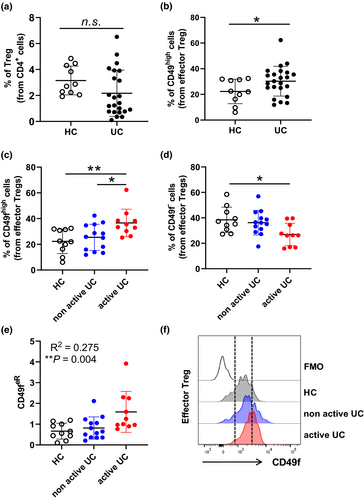
In summary, our data support the notion that active UC is associated with increased CD49f expression on circulating Treg and that the assessment of CD49f ratios within the effector Treg compartment may be a useful predictor of disease activity.
Discussion
Through a combination of in vitro functional studies, quantitative proteomics, transcriptome deep sequencing and phenotypic analyses, we identified unexplored subsets of Treg defined by CD49f expression. This finding addresses a gap in understanding Treg immunomodulatory function in homeostasis and human diseases. The CD49f− Treg subset exhibits a unique phenotypic profile with significantly increased suppressive capacity. In contrast, the frequencies of Th17-like CD49fhigh Treg correlated with the activity of UC, suggesting that subset exclusion based on CD49fhigh expression on Treg may constitute a promising strategy to maximise the efficacy and safety of Treg-based immunotherapy for treating patients with IBD. Understanding the pathogenic role of CD49fhigh Treg in inflammatory disorders may provide insight into the drivers of maladaptive inflammation.
CD49f plays an important and conserved role in stem cell biology.23 It belongs to the integrin family of receptors, which are structurally characterised as transmembrane adhesion receptors that mediate cell–cell and cell–extracellular matrix adhesion and induces bidirectional signalling across the cell membrane that regulates proliferation, activation, migration and homeostasis.23 CD49f often dimerises with β1 and β6 integrins to form heterodimers including α6β1 and α6β4 that act as primary receptors for laminins present in their niche.24 Accumulation of evidence from human and mouse models shows that defects in integrin expression or unintentional inflammation against healthy host tissue result in serious immunodeficiency and many autoimmune conditions.25, 26 Accordingly, integrins such as CD49b and CD49d have been recently described to modulate various aspects of Treg biology.17, 18, 27, 28 Human CD49d− Treg have been reported to present higher immunosuppressive function than their CD49d+ counterparts.18 This finding is supported by more recent evidence that CD49ddim/− expression enriches for a subset of cells with suppressive capacity within the CD8+CD122+ population of effector T cells.29 In contrast, the expression of CD49b on mature Treg that survey the skin and vascular tissues resulted in superior suppressive capacity and decreased disease severity in a mouse model of T cell-induced arthritis, partially dependent on IL-10 secretion.28, 30 Unlike CD49d and CD49b, information on the Treg immune-modulating capacity by CD49f is scarce.
In this study, we show that CD49f expression defines a subset of Treg with impaired suppressive capacity and decreased expression of the functional Treg markers CTLA4, CD39 and FoxP3. FoxP3 is not only required for differentiation of Treg towards a suppressive phenotype but also a prerequisite for stabilising the Treg lineage.31 Similarly, high expression of CD39 on human Treg drives cell stability and function under inflammatory conditions through the conversion of ATP into adenosine and AMP,32 whereas deficiency of CTLA4 in Treg is associated with the development of spontaneous systemic lymphoproliferation and fatal T cell-mediated autoimmune disease.33 Interestingly, although CD49f expression on resting and effector Treg appears similar, CD49f only impacted the expression of functional markers on effector Treg, indicating that rather than being a marker for effector versus resting Treg, CD49f is associated with Treg suppressive function and pro-inflammatory profile in the effector compartment.
UC is a chronic autoimmune disease characterised by infiltration of inflammatory cells into the lamina propria of the intestinal tract. Various subsets of intestinal lamina propria T cells are believed to traffic, via the systemic circulation, from gut-associated lymphoid tissue.34 Even though it is assumed that the absence of Treg leads to IBD in both human and mice models,4-6 there is little evidence to suggest that patients with IBD simply lack Treg in the circulation and/or the affected tissues.35-37 In our cohort, patients with UC had a lower number of circulating Treg, but this was not statistically significant. However, CD49fhigh effector Treg were significantly increased in these patients in comparison with healthy controls, and an increased ratio of CD49fhigh/CD49f− effector Treg (CD49feR) was an indicator of disease activity. Thus, the role of Treg in IBD requires a more nuanced approach than simple enumeration of T cells bearing classic Treg markers.
It is thought that the development of UC is underpinned by an imbalance between Th17 and Treg cells. UC is associated with a sequestration of immune cells within the gut mucosa, where a pro-inflammatory cytokine environment restricts Treg activity and promotes the continual differentiation and development of a dysregulated Th17 response.38 The generation of Th17 cells requires the expression of RORγt, originally defined as a thymic-specific isoform of RORγ.39 In patients with UC presenting moderate and severe disease, the inflammatory response is positively correlated with IL-17 expression in colonic specimens.11
Effector Th17-like Treg have also been described in the IBD scenario. Ex vivo secretion of IL-17A and constitutive expression of the chemokine receptor CCR6 along with RORγt in human effector Treg suggest that these cells are damaging entities.40 Besides its classical role in regulating Th17 cell migration, CCR6 expression on Treg is an important mediator of their recruitment into inflammatory tissues.41 Thus, CD49fhigh effector Treg expressing increased level of CCR6 at both the transcriptional and protein levels are likely to present higher adhesion and migration within the extracellular matrix of the intestinal lamina propria and exacerbates IBD. Corroborating with this hypothesis, a previous study demonstrated downregulation of CD49f on the surface of both CD4+ and CD8+ conventional T cells following migration into the inflamed intestinal lamina propria of patients with IBD.19
Because of the high Treg plasticity, the ontogenesis of Th17-like Treg is still under debate and some authors suggested that they might represent a transient stage of progenitor cells that can convert into either FoxP3+ Treg or Th17 cells under certain inflammatory conditions including UC.13, 14, 42 Interestingly, CD49f may be the only marker commonly found in more than thirty different populations of stem cells, including some hematopoietic stem cells.43 Thus, it is possible that CD49f expression is associated with the preservation of Th17-like Treg cells in a progenitor stage, thus contributing to a pro-inflammatory milieu that leads to the development of IBD. The mechanisms that drive CD49f expression on Treg still remain to be elucidated, and it is possible that the expression is underpinned by TGF-β-related cytokines. While TGF-β signalling is important for development and maintenance of Treg, it also upregulates the expression of several types of integrin receptors.43 Our previous findings demonstrate that CD49f (integrin alpha 6) expression on thymic epithelial progenitor cells is modulated by the members of the TGF-β superfamily of proteins.44
Here, we propose a model where CD49f modulates Treg cell function and differentiation in humans. While the absence of CD49f renders Treg more suppressive and likely to play a vital role in immune homeostasis under normal physiological conditions, high expression of CD49f seems to contribute at least in part to the development of pro-inflammatory effector Treg that correlate with disease activity in UC. Our results highlight the importance of CD49f in modulating Treg mechanisms that guide homeostasis in health and dysfunction in disease.
METHODS
Specimens from patients with UC and healthy donors
Cryopreserved PBMC from 23 patients with UC part of the Mater Hospital IBD biobank, Brisbane, Australia, were assessed by flow cytometric analysis (Table 1). Age- and gender-matched PBMC from healthy donors were included as controls. Blood samples were collected as part of the Mater IBD biobank (Mater HREC approval AM/MML/24730). PBMC from patients receiving anti-TNF therapy were excluded from our analysis because of direct effect of TNF-α on Treg.45 Before staining, cryopreserved samples were thawed and incubated in RPMI 1640 containing 10 µg mL−1 DNAse I (Roche, Basel, Switzerland) at 37°C for 1 h to prevent cell clumping and debris. PBMC from healthy controls were freshly isolated from volunteers at QIMR Berghofer for transcriptome, proteomic and functional studies. Ethics approval was obtained from the human research ethics committee QIMR Berghofer, Brisbane, Queensland, Australia (HREC #P2058). In all cases, PBMC were isolated using a Ficoll-Paque Plus (Merck, Kenilworth, New Jersey, USA) density gradient centrifugation from blood and a written informed consent was obtained from volunteers.
Isolation of human T cells
CD4+ T cells were separated from PBMC from healthy controls using pan T-cell isolation kit and magnetic-activated cell sorting (Miltenyi Biotec, Bergisch Gladbach, North Rhine-Westphalia, Germany). Enriched T cells were subsequently stained with CD25-PE mAb and CD25+ T cells positively selected using anti-PE magnetic beads and MACS (Miltenyi Biotec, Bergisch Gladbach, North Rhine-Westphalia, Germany). To obtain high purity Treg, CD25highCD127− T cells were subsequently sorted in FACSAria III (BD Biosciences, Franklin Lakes, New Jersey, USA). The purity of the sorted Treg subsets was assessed by staining the cells with FoxP3-APC mAb. CD49f- and CD49fhigh cells were further separated from the total fraction of Treg by FACS-based sorting based on CD49f FMO controls. Over 95% of purity was detected in all cases analysed. The remaining CD25− fraction was assessed as conv CD4+ populations.
Proteomic sample preparation and LC-MS/MS analysis
Approximately 106 cells were lysed in SDS-containing buffer for proteomic analysis. Trypsin digestion using the protein co-precipitation method with trypsin and peptide desalting was performed as described.46 Based on micro-BCA assay protein quantification, 0.9 μg of tryptic peptide samples and label-free shotgun proteomic data were obtained on Orbitrap Fusion™ Tribrid™ mass spectrometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), inline coupled to nanoACQUITY ultra-performance liquid chromatography system (Waters, USA), using Symmetry C18, 2G, VM trap columns (100 Å, 5 μm particle size, 180 μm × 20 mm) and BEH C18 analytical columns (130 Å, 1.7 μm particle size, 75 μm × 200 mm) at a flow rate of 3 μL min−1 over 175 min. The mobile phase consisted of solvent A (0.1% formic acid) and solvent B (100% acetonitrile/0.1% formic acid). Three consecutive linear gradients were used for peptide elution: 5%–9% of solvent B between 3 and 10 min, 9%–26% of solvent B from 10 to 120 min and 26–40% of solvent B from 120 to 145 min. Column cleaning and equilibration was achieved with gradient from 40 to 80% of solvent B at 145–152 min, holding at 80% until 157 min and then at 1% until 160 min. EASY-Max NG™ ion source (Thermo Fisher Scientific, Waltham, Massachusetts, USA) was applied at 1900V and 285°C to ionise the eluted peptides. Xcalibur software version 3.0.63 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at ‘top speed’ mode allowed the automatic selection of positively charged peptides (+2 to +7) in a 2-s cycle time.
Identification of signature proteome of Treg cells
Acquired proteomic data were searched using MaxQuant (Release 1.5.8.3) software47 against UniProt human reviewed proteome database containing 20, 242 entries (downloaded on 25th October 2017). Carbamidomethylation was assessed as the fixed modification, while oxidation and N-terminal acetylation were considered as the variable modifications. maxLFQ included in MaxQuant software was used to obtain the normalised label-free peptide and protein intensity data. Proteins quantified by at least 2 unique or razor peptides at m-score of > 5 and on > 50% of the samples were selected for further analysis. Missing protein intensity values of the selected proteins were imputed using maximum-likelihood estimate (R package), and differential expression analysis was performed using multiple t-test with FDR determination by two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli. In the DE analysis, protein expression data of Treg cells were compared with conv CD4+ cells to obtain the log2FC and the statistical significance. DE proteins were defined as log2FC ≥ 1 or ≤ −1 at FDR value of < 0.05. These proteins were further analysed using IPA core analysis to identify proteins related to cell surface. Each of the proteins annotated as plasma membrane in subcellular localisation were individually searched to identify potentially uncharacterised surface proteins in circulating human Treg cells.
Antibodies and flow cytometry
Fluorescence dye-labelled antibodies specific for human CD3, CD4, CD25, CD127, CD45RA, CD49f, IL-17A, IFNγ, CTLA4, CD39, CCR6 and FoxP3 were used for flow cytometric analysis and sorting (Supplementary table 4). Intranuclear staining for FoxP3 was achieved using FoxP3 Fix/Perm Buffer Set for nuclear staining (BioLegend, San Diego, California, USA). Intracellular IL-17A staining was performed using Cytofix/Cytoperm buffer for intracellular staining (BD Biosciences, Franklin Lakes, New Jersey, USA). Viability of cells was defined by LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Samples were acquired by a BD Fortessa multiparametric flow cytometer (BD Biosciences, Franklin Lakes, New Jersey, USA).
In vitro T-cell suppression assay
Suppression of conv CD4+ cell proliferation by Treg cells was assessed based on an assay previously optimised for small number of cells.48 Treg cells were sorted as CD49fhigh or CD49f− populations. The assay was carried out in a 96-well round-bottom plate where 25 000 of conv CD4+ cells previously stained with cell trace violet (CTV, Thermo Fisher Scientific, Waltham, Massachusetts, USA) were co-cultured with Treg cells at Treg: conv CD4+ cell ratio ranging from 1:1 to 1:16. Between three and four technical replicates were used in each condition and analysed. To stimulate proliferation, conv CD4+ cells were activated with 1 μg mL−1 of soluble anti-human CD3/OKT3 mAb (Sigma-Aldrich, St. Louis, Missouri, USA) in the presence of irradiated allogenic PBMC (˜50,000) for five days. Conv CD4+ cells stimulated without Treg cells were also included in the assay to monitor basal T-cell proliferation. Proliferation of conv CD4+ cell was assessed with FlowJo software version 10 (TreeStar, Ashland, Oregon, USA). Unstimulated conv CD4+ cells were used to establish the gating for CTV−/low cells. The percentage of Treg-mediated suppression was calculated as 100−((proliferated cells with Treg/proliferated cells with no Treg) × 100).
Assessment of cytokine production by Treg
IL-17A and IFNγ cytokines production by CD49f− and CD49fhigh Treg was assessed ex vivo using intracellular staining. Briefly, 125 000 Tregs were plated per well on a 96-well round-bottom plate in the presence of 500 IU mL−1 of human recombinant IL-2 (Novartis, Basel, Switzerland) and CD3/CD28 Dynabeads, using three Dynabeads per one Treg cell (Thermo Fisher Scientific, Waltham, Massachusetts, USA) for activation. Cells were incubated overnight at 37°C. Brefeldin A was added at 10 μg mL−1 (BD Biosciences, Franklin Lakes, New Jersey, USA) for the last 4 h of incubation. After this period, cells were stained with IL-17A and IFNγ mAbs for flow cytometric analysis.
RNA library preparation
RNA was purified from approximately 25 000 of FACS-isolated CD49fhigh and CD49f− Treg using the Arcturus PicoPure Isolation Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). RNA integrity was confirmed on the Agilent 2100 Bioanalyser using the Total RNA Pico Kit (Agilent Technologies, Santa Clara, California, USA). Oligo d(T) captured mRNA was processed for next-generation sequencing (NGS) with the NEB Next Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts, USA). Quality of purified RNA was assessed on the Agilent 2100 Bioanalyser using the High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, California, USA). RNA quantification was based on the Qubit DNA HS Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Final libraries were pooled and sequenced using a High output single-end 75 cycle (version 2) sequencing kit and the Illumina Nextseq 550 platform (Illumina, San Diego, California, USA).
Bioinformatics analysis of RNA-sequencing data
Reads were trimmed for adapter sequences using Cutadapt (version 1.11)49 and aligned by STAR50 (version 2.5.2a) to the GRCh37 assembly using the gene, transcript, and exon features of Ensembl (release 70) gene model. Expression was estimated through RSEM (version 1.2.30).51 Transcripts with zero read counts across all samples were removed prior to analysis. Normalisation of read counts was performed by dividing million reads mapped to generate counts per million (CPM), followed by the trimmed mean of M-values (TMM) method from the edgeR package (version 3.32.0). For the differential expression analyses, the glmFit function was adapted to fit a negative binomial generalised log-linear model to the read counts for each transcript. Using the glmLRT function, we conducted transcript wise likelihood ratio tests for each comparison. Log2 transformed, normalised read counts were used for heatmaps and PCA. Hierarchical clustering of genes and samples in the heatmaps was performed with average linkage clustering on the 1- Spearman correlation coefficient dissimilarity matrix of all DE transcripts with a FDR < 0.05 or as stated in text.
Gene set enrichment and pathway analysis
Gene set enrichment analysis was performed using GSEA (Broad Institute, Cambridge, Massachusetts, USA).52 P-values were generated form 1000 gene set permutations, excluding gene sets with more than 3000 or less than 5 genes against custom-made gene sets (GSE107011)53, 54 and Broads Hallmark database. In addition, IPA was used with the default settings to identity canonical pathways of DE gene transcripts (FDR < 0.05).
Quantification and statistical analysis
All values are expressed as mean ± SD, unless otherwise specified. Statistical analyses were performed using GraphPad Prism v7.02 (GraphPad, San Diego, California, USA), with the appropriate tests utilised. A P-value < 0.05 was considered statistically significant.
Acknowledgments
We thank the volunteers for donating blood samples for this study. HW was supported by an International Postgraduate Research Scholarship, The University of Queensland, Brisbane, Australia, and PhD Top-up Scholarship, QIMR Berghofer Medical Research Institute, Brisbane, Australia. JJM is supported by a NHMRC CDF Level 2 Fellowship (1131732). The authors thank the expertise of staff from the Translational Research Institute FlowCore facility and the QIMR Berghofer Analytical facility.
Conflict of interests
The authors declare no competing interests.
Author contributions
Harshi Weerakoon: Data curation; Formal analysis; Investigation; Methodology; Writing-original draft. Jasmin Straube: Data curation; Formal analysis; Software; Writing-review & editing. Katie Lineburg: Formal analysis; Investigation. Leanne Cooper: Investigation. Steven Lane: Investigation; Supervision. Corey Smith: Supervision; Writing-review & editing. Saleh Alabbas: Investigation; Resources. Jakob Begun: Resources; Supervision; Writing-review & editing. John Miles: Methodology; Resources; Supervision; Writing-review & editing. Michelle M Hill: Conceptualization; Methodology; Resources; Supervision; Writing-original draft. Ailin Lepletier: Conceptualization; Formal analysis; Investigation; Methodology; Supervision; Writing-original draft.
Data availability statement
RNA-Seq data reported in this paper are available on the GEO repository with accession number GSE161077. The mass spectrometry proteomics data have been deposited on the ProteomeXchange Consortium via the PRIDE55 partner repository with the data set identifier PXD022095.




