Unlocking Biomolecular Activity through Pd-Catalyzed Azides Reduction†
Fang Fu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorWei Xiong
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorXinyan Xu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorYongjie Liu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorMing Li
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorQianqian Qi
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorXingyu Liu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorYuanyuan Zhang
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorCorresponding Author
Tian Tian
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
E-mail: [email protected]Search for more papers by this authorXiang Zhou
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorFang Fu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorWei Xiong
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorXinyan Xu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
These authors contributed equally to this work.
Search for more papers by this authorYongjie Liu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorMing Li
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorQianqian Qi
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorXingyu Liu
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorYuanyuan Zhang
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorCorresponding Author
Tian Tian
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
E-mail: [email protected]Search for more papers by this authorXiang Zhou
Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorDedicated to the Special Issue of Emerging Investigators in 2024.
Comprehensive Summary
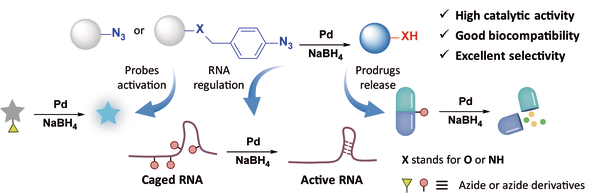
Pd-mediated bioorthogonal cleavage reactions have been extensively utilized in the activation of prodrug molecules, precise regulation of protein function, and cellular engineering. However, the availability of cleavable "caging" groups is quite limited, and their application in nucleic acid modification has seldom been reported. Herein, we introduce a method based on Pd-catalyzed reduction amination of azides as a decaging strategy to activate the activity of biomolecules. We designed modifications on the bioactive sites with azides or their derivatives to mask the related biological function, followed by the release of biological activity through Pd-catalyzed NaBH4 reduction amination reaction. This study has demonstrated that the strategy can effectively be used to activate bioactive molecules such as fluorescent probes, prodrugs, and to regulate the biological function of RNA, including reverse transcription extension, binding to ligands, and cleavage activity of the CRISPR-Cas system. All results confirm that this strategy provides an efficient and controllable "OFF to ON" biological switch, capable of achieving significant regulatory effects substoichiometrically, and is expected to be extended to other biological applications.
Supporting Information
| Filename | Description |
|---|---|
| cjoc202400254-sup-0001-supinfo.pdfPDF document, 3 MB |
Appendix S1: Supporting information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1 Fedeli, S.; Im, J.; Gopalakrishnan, S.; Elia, J. L.; Gupta, A.; Kim, D.; Rotello, V. M. Nanomaterial-based bioorthogonal nanozymes for biological applications. Chem. Soc. Rev. 2021, 50, 13467–13480.
- 2 Li, J.; Chen, P. R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 2016, 12, 129–137.
- 3 Li, Y.; Fu, H. Bioorthogonal Ligations and Cleavages in Chemical Biology. ChemistryOpen 2020, 9, 835–853.
- 4 Bai, Y.; Chen, J.; Zimmerman, S. C. Designed transition metal catalysts for intracellular organic synthesis. Chem. Soc. Rev. 2018, 47, 1811–1821.
- 5 Deb, T.; Tu, J.; Franzini, R. M. Mechanisms and Substituent Effects of Metal-Free Bioorthogonal Reactions. Chem. Rev. 2021, 121, 6850–6914.
- 6 Fairbanks, B. D.; Macdougall, L. J.; Mavila, S.; Sinha, J.; Kirkpatrick, B. E.; Anseth, K. S.; Bowman, C. N. Photoclick Chemistry: A Bright Idea. Chem. Rev. 2021, 121, 6915–6990.
- 7 Kumar, G. S.; Lin, Q. Light-Triggered Click Chemistry. Chem. Rev. 2021, 121, 6991–7031.
- 8 Bird, R. E.; Lemmel, S. A.; Yu, X.; Zhou, Q. A. Bioorthogonal Chemistry and Its Applications. Bioconjugate Chem. 2021, 32, 2457–2479.
- 9 Liang, T.; Chen, Z.; Li, H.; Gu, Z. Bioorthogonal catalysis for biomedical applications. Trends Chem. 2022, 4, 157–168.
- 10 Wang, J.; Wang, X.; Fan, X.; Chen, P. R. Unleashing the Power of Bond Cleavage Chemistry in Living Systems. ACS Cent. Sci. 2021, 7, 929–943.
- 11 Soldevila-Barreda, J. J.; Metzler-Nolte, N. Intracellular Catalysis with Selected Metal Complexes and Metallic Nanoparticles: Advances toward the Development of Catalytic Metallodrugs. Chem. Rev. 2019, 119, 829–869.
- 12 Wang, J.; Zheng, S.; Liu, Y.; Zhang, Z.; Lin, Z.; Li, J.; Zhang, G.; Wang, X.; Li, J.; Chen, P. R. Palladium-triggered chemical rescue of intracellular proteins via genetically encoded allene-caged tyrosine. J. Am. Chem. Soc. 2016, 138, 15118–15121.
- 13 Yusop, R. M.; Unciti-Broceta, A.; Johansson, E. M. V.; Sánchez-Martín, R. M.; Bradley, M. Palladium-mediated intracellular chemistry. Nat. Chem. 2011, 3, 239–243.
- 14 Sancho-Albero, M.; Rubio-Ruiz, B.; Pérez-López, A. M.; Sebastián, V.; Martín-Duque, P.; Arruebo, M.; Santamaría, J.; Unciti-Broceta, A. Cancer-derived exosomes loaded with ultrathin palladium nanosheets for targeted bioorthogonal catalysis. Nat. Catal. 2019, 2, 864–872.
- 15 Martínez-Calvo, M.; Couceiro, J. R.; Destito, P.; Rodríguez, J.; Mosquera, J.; Mascareñas, J. L. Intracellular deprotection reactions mediated by palladium complexes equipped with designed phosphine ligands. ACS Catal. 2018, 8, 6055–6061.
- 16 Miller, M. A.; Askevold, B.; Mikula, H.; Kohler, R. H.; Pirovich, D.; Weissleder, R. Nano-palladium is a cellular catalyst for in vivo chemistry. Nat. Commun. 2017, 8, 15906.
- 17 Weiss, J. T.; Dawson, J. C.; Macleod, K. G.; Rybski, W.; Fraser, C.; Torres-Sánchez, C.; Patton, E. E.; Bradley, M.; Carragher, N. O.; Unciti-Broceta, A. Extracellular palladium-catalysed dealkylation of 5-fluoro-1-propargyl-uracil as a bioorthogonally activated prodrug approach. Nat. Commun. 2014, 5, 3277.
- 18 Li, J.; Yu, J.; Zhao, J.; Wang, J.; Zheng, S.; Lin, S.; Chen, L.; Yang, M.; Jia, S.; Zhang, X.; Chen, P. R. Palladium-triggered deprotection chemistry for protein activation in living cells. Nat. Chem. 2014, 6, 352–361.
- 19 Konč, J.; Sabatino, V.; Jiménez-Moreno, E.; Latocheski, E.; Pérez, L. R.; Day, J.; Domingos, J. B.; Bernardes, G. J. L. Controlled in-cell generation of active palladium(0) species for bioorthogonal decaging. Angew. Chem. Int. Ed. 2022, 61, e202113519.
- 20 Chen, Z.; Li, H.; Bian, Y.; Wang, Z.; Chen, G.; Zhang, X.; Miao, Y.; Wen, D.; Wang, J.; Wan, G.; Zeng, Y.; Abdou, P.; Fang, J.; Li, S.; Sun, C.-J.; Gu, Z. Bioorthogonal catalytic patch. Nat. Nanotechnol. 2021, 16, 933–941.
- 21 Liu, J.; Liu, X.; Liu, Q.; Cao, J.; Lv, X.; Wang, S.; Tian, T.; Zhou, X.; Deng, H. Mesoporous Metal–Organic Frameworks for Catalytic RNA Deprotection and Activation. Angew. Chem. Int. Ed. 2023, 62, e202302649.
- 22 Dal Forno, G. M.; Latocheski, E.; Beatriz Machado, A.; Becher, J.; Dunsmore, L.; St. John, A. L.; Oliveira, B. L.; Navo, C. D.; Jiménez-Osés, G.; Fior, R.; Domingos, J. B.; Bernardes, G. J. L. Expanding Transition Metal-Mediated Bioorthogonal Decaging to Include C–C Bond Cleavage Reactions. J. Am. Chem. Soc. 2023, 145, 10790–10799.
- 23 Pérez-López, A. M.; Rubio-Ruiz, B.; Sebastián, V.; Hamilton, L.; Adam, C.; Bray, T. L.; Irusta, S.; Brennan, P. M.; Lloyd-Jones, G. C.; Sieger, D.; Santamaría, J.; Unciti-Broceta, A. Gold-Triggered Uncaging Chemistry in Living Systems. Angew. Chem. Int. Ed. 2017, 56, 12548–12552.
- 24 Tsubokura, K.; Vong, K. K. H.; Pradipta, A. R.; Ogura, A.; Urano, S.; Tahara, T.; Nozaki, S.; Onoe, H.; Nakao, Y.; Sibgatullina, R.; Kurbangalieva, A.; Watanabe, Y.; Tanaka, K. In Vivo Gold Complex Catalysis within Live Mice. Angew. Chem. Int. Ed. 2017, 56, 3579–3584.
- 25 Jbara, M.; Eid, E.; Brik, A. Gold(I)-Mediated Decaging or Cleavage of Propargylated Peptide Bond in Aqueous Conditions for Protein Synthesis and Manipulation. J. Am. Chem. Soc. 2020, 142, 8203–8210.
- 26 Vong, K.; Yamamoto, T.; Chang, T.-c.; Tanaka, K. Bioorthogonal release of anticancer drugs via gold-triggered 2-alkynylbenzamide cyclization. Chem. Sci. 2020, 11, 10928–10933.
- 27 Sabatino, V.; Rebelein, J. G.; Ward, T. R. “Close-to-Release”: Spontaneous Bioorthogonal Uncaging Resulting from Ring-Closing Metathesis. J. Am. Chem. Soc. 2019, 141, 17048–17052.
- 28 Sasmal, P. K.; Carregal-Romero, S.; Parak, W. J.; Meggers, E. Light-Triggered Ruthenium-Catalyzed Allylcarbamate Cleavage in Biological Environments. Organometallics 2012, 31, 5968–5970.
- 29 Völker, T.; Dempwolff, F.; Graumann, P. L.; Meggers, E. Progress towards Bioorthogonal Catalysis with Organometallic Compounds. Angew. Chem. Int. Ed. 2014, 53, 10536–10540.
- 30 Tomás-Gamasa, M.; Martínez-Calvo, M.; Couceiro, J. R.; Mascareñas, J. L. Transition metal catalysis in the mitochondria of living cells. Nat. Commun. 2016, 7, 12538.
- 31 Vidal, C.; Tomás-Gamasa, M.; Destito, P.; López, F.; Mascareñas, J. L. Concurrent and orthogonal gold(I) and ruthenium(II) catalysis inside living cells. Nat. Commun. 2018, 9, 1913.
- 32 Singh, N.; Gupta, A.; Prasad, P.; Mahawar, P.; Gupta, S.; Sasmal, P. K. Iridium-Triggered Allylcarbamate Uncaging in Living Cells. Inorg. Chem. 2021, 60, 12644–12650.
- 33 Wang, X.; Liu, Y.; Fan, X.; Wang, J.; Ngai, W. S. C.; Zhang, H.; Li, J.; Zhang, G.; Lin, J.; Chen, P. R. Copper-triggered bioorthogonal cleavage reactions for reversible protein and cell surface modifications. J. Am. Chem. Soc. 2019, 141, 17133–17141.
- 34 Oliveira, B. L.; Stenton, B. J.; Unnikrishnan, V. B.; de Almeida, C. R.; Conde, J.; Negrão, M.; Schneider, F. S. S.; Cordeiro, C.; Ferreira, M. G.; Caramori, G. F.; Domingos, J. B.; Fior, R.; Bernardes, G. J. L. Platinum-triggered bond-cleavage of pentynoyl amide and N-propargyl handles for drug-activation. J. Am. Chem. Soc. 2020, 142, 10869–10880.
- 35 Cao-Milán, R.; Gopalakrishnan, S.; He, L. D.; Huang, R.; Wang, L.-S.; Castellanos, L.; Luther, D. C.; Landis, R. F.; Makabenta, J. M. V.; Li, C.-H.; Zhang, X.; Scaletti, F.; Vachet, R. W.; Rotello, V. M. Thermally Gated Bio-orthogonal Nanozymes with Supramolecularly Confined Porphyrin Catalysts for Antimicrobial Uses. Chem 2020, 6, 1113–1124.
- 36 Huang, C.; Zhao, C.; Deng, Q.; Zhang, H.; Yu, D.; Ren, J.; Qu, X. Hydrogen-bonded organic framework-based bioorthogonal catalysis prevents drug metabolic inactivation. Nat. Catal. 2023, 6, 729–739.
- 37 Yin; Liebscher, J. Carbon−carbon coupling reactions catalyzed by heterogeneous palladium catalysts. Chem. Rev. 2007, 107, 133–173.
- 38 Li, H.; Johansson Seechurn, C. C. C.; Colacot, T. J. Development of preformed Pd catalysts for cross-coupling reactions, beyond the 2010 Nobel Prize. ACS Catal. 2012, 2, 1147–1164.
- 39 Gildner, P. G.; Colacot, T. J. Reactions of the 21st Century: Two Decades of Innovative Catalyst Design for Palladium-Catalyzed Cross-Couplings. Organometallics 2015, 34, 5497–5508.
- 40 George, J. T.; Srivatsan, S. G. Vinyluridine as a versatile chemoselective handle for the post-transcriptional chemical functionalization of RNA. Bioconjugate Chem. 2017, 28, 1529–1536.
- 41 Walunj, M. B.; Tanpure, A. A.; Srivatsan, S. G. Post-transcriptional labeling by using Suzuki–Miyaura cross-coupling generates functional RNA probes. Nucleic Acids Res. 2018, 46, e65.
- 42 Li, J.; Lin, S.; Wang, J.; Jia, S.; Yang, M.; Hao, Z.; Zhang, X.; Chen, P. R. Ligand-free palladium-mediated site-specific protein labeling inside gram-negative bacterial pathogens. J. Am. Chem. Soc. 2013, 135, 7330–7338.
- 43 Simmons, R. L.; Yu, R. T.; Myers, A. G. Storable arylpalladium(II) reagents for alkene labeling in aqueous media. J. Am. Chem. Soc. 2011, 133, 15870–15873.
- 44 Jbara, M.; Maity, S. K.; Brik, A. Palladium in the Chemical Synthesis and Modification of Proteins. Angew. Chem. Int. Ed. 2017, 56, 10644–10655.
- 45 Hirschbiegel, C.-M.; Zhang, X.; Huang, R.; Cicek, Y. A.; Fedeli, S.; Rotello, V. M. Inorganic nanoparticles as scaffolds for bioorthogonal catalysts. Adv. Drug Delivery Rev. 2023, 195, 114730.
- 46 Latocheski, E.; Dal Forno, G. M.; Ferreira, T. M.; Oliveira, B. L.; Bernardes, G. J. L.; Domingos, J. B. Mechanistic insights into transition metal-mediated bioorthogonal uncaging reactions. Chem. Soc. Rev. 2020, 49, 7710–7729.
- 47 Liu, S.; Edgar, K. J. Staudinger Reactions for Selective Functionalization of Polysaccharides: A Review. Biomacromolecules 2015, 16, 2556–2571.
- 48 Nepomniaschiy, N.; Grimminger, V.; Cohen, A.; DiGiovanni, S.; Lashuel, H. A.; Brik, A. Switch Peptide via Staudinger Reaction. Org. Lett. 2008, 10, 5243–5246.
- 49 Luo, J.; Liu, Q.; Morihiro, K.; Deiters, A. Small-molecule control of protein function through Staudinger reduction. Nat. Chem. 2016, 8, 1027–1034.
- 50 Liu, G.; Wold, E. A.; Zhou, J. Applications of Bioorthogonal Chemistry in Tumor-Targeted Drug Discovery. Curr. Top. Med. Chem. 2019, 19, 892–897.
- 51 Lei, H.; Zeng, T.; Ye, X.; Fan, R.; Xiong, W.; Tian, T.; Zhou, X. Chemical Control of CRISPR Gene Editing via Conditional Diacylation Crosslinking of Guide RNAs. Adv. Sci. 2023, 10, 2206433.
- 52 Chen, Y.; Yang, L.; Zhang, X.; Deng, S.; You, L.; Liu, Y. Copper-Catalyzed Reduction of Azides with Hydrosilanes. ChemistrySelect 2018, 3, 96–99.
- 53 Liu, L.; Wang, Q.; Liu, Y.; Zhang, X.; Lu, D.; Deng, S.; Gao, Y.; Chen, Y. Copper catalyzed reduction of azides with diboron under mild conditions. Tetrahedron Lett. 2020, 61, 151702.
- 54 Ahammed, S.; Saha, A.; Ranu, B. C. Hydrogenation of Azides over Copper Nanoparticle Surface Using Ammonium Formate in Water. J. Org. Chem. 2011, 76, 7235–7239.
- 55 Pimpasri, C.; White, A. J. P.; Díez-González, S. User-friendly copper-catalysed reduction of azides to amines. Asian J. Org. Chem. 2020, 9, 399–403.
- 56 Sasmal, P. K.; Carregal-Romero, S.; Han, A. A.; Streu, C. N.; Lin, Z.; Namikawa, K.; Elliott, S. L.; Köster, R. W.; Parak, W. J.; Meggers, E. Catalytic azide reduction in biological environments. ChemBioChem 2012, 13, 1116–1120.
- 57 Udumula, V.; Nazari, S. H.; Burt, S. R.; Alfindee, M. N.; Michaelis, D. J. Chemo- and site-selective alkyl and aryl azide reductions with heterogeneous nanoparticle catalysts. ACS Catal. 2016, 6, 4423–4427.
- 58 Cantopcu, E.; Aydinli, E.; Goksu, H. Homogeneous catalyst containing Pd in the reduction of aryl azides to primary amines. J. Chem. Sci. 2022, 134, 41.
- 59 He, X.; Chen, X.; Wang, Y. Mass Spectrometry for Assessing Protein–Nucleic Acid Interactions. Anal. Chem. 2023, 95, 115–127.
- 60 Ramanathan, M.; Porter, D. F.; Khavari, P. A. Methods to study RNA–protein interactions. Nat. Methods 2019, 16, 225–234.
- 61 Huang, K.; Chen, X.; Li, C.; Song, Q.; Li, H.; Zhu, L.; Yang, Y.; Ren, A. Structure-based investigation of fluorogenic Pepper aptamer. Nat. Chem. Biol. 2021, 17, 1289–1295.
- 62 Zhang, D.; Liu, L.; Jin, S.; Tota, E.; Li, Z.; Piao, X.; Zhang, X.; Fu, X.-D.; Devaraj, N. K. Site-Specific and Enzymatic Cross-Linking of sgRNA Enables Wavelength-Selectable Photoactivated Control of CRISPR Gene Editing. J. Am. Chem. Soc. 2022, 144, 4487–4495.
- 63 Zhang, F.; Wen, Y.; Guo, X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014, 23, R40–R46.
- 64 Liu, X.-Y.; Xiong, W.; Qi, Q.-Q.; Ji, H.-M.; Zhang, Y.-T.; Lei, H.-J.; Liu, J.; Yin, P.; Tian, T.; Zhou, X. A chemical CRISPR off switch efficiently controls gene editing. Cell Rep. Phys. Sci. 2022, 3, 100956.
- 65 Wang, S.; Wei, L.; Wang, J.-Q.; Ji, H.; Xiong, W.; Liu, J.; Yin, P.; Tian, T.; Zhou, X. Light-Driven Activation of RNA-Guided Nucleic Acid Cleavage. ACS Chem. Biol. 2020, 15, 1455–1463.
- 66 Jain, P. K.; Ramanan, V.; Schepers, A. G.; Dalvie, N. S.; Panda, A.; Fleming, H. E.; Bhatia, S. N. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angew. Chem. Int. Ed. 2016, 55, 12440–12444.
- 67 East-Seletsky, A.; O'Connell, M. R.; Knight, S. C.; Burstein, D.; Cate, J. H. D.; Tjian, R.; Doudna, J. A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273.
- 68 Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726.e10.
- 69 Cox, D. B. T.; Gootenberg, J. S.; Abudayyeh, O. O.; Franklin, B.; Kellner, M. J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027.




