Intracellular RNA Labeling Technologies for the Analysis of RNA Biology†
Ruiqi Zhao
Department of Clinical Laboratory, Center for Gene Diagnosis, Department of Neurosurgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
These authors contributed equally.
Search for more papers by this authorXin Fang
College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, 430072 China
These authors contributed equally.
Search for more papers by this authorCorresponding Author
Xiaocheng Weng
Department of Clinical Laboratory, Center for Gene Diagnosis, Department of Neurosurgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, 430072 China
*E-mail: [email protected]Search for more papers by this authorRuiqi Zhao
Department of Clinical Laboratory, Center for Gene Diagnosis, Department of Neurosurgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
These authors contributed equally.
Search for more papers by this authorXin Fang
College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, 430072 China
These authors contributed equally.
Search for more papers by this authorCorresponding Author
Xiaocheng Weng
Department of Clinical Laboratory, Center for Gene Diagnosis, Department of Neurosurgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, 430072 China
*E-mail: [email protected]Search for more papers by this authorDedicated to the 130th Anniversary of Wuhan University.
Comprehensive Summary
As a cornerstone of the central dogma of molecular biology, RNA plays vital roles in living organisms. Over the past few decades, many RNA labeling technologies have been developed to elucidate the biological function of RNA. These technologies have significantly advanced our understanding of RNA secondary structure, localization, and turnover. Additionally, taking advantage of these innovative RNA labeling approaches, plenty of tool kits have been devised for the regulation of RNA-related biological process, such as gene expression and gene editing. In this review, we primarily focus on an array of intracellular RNA labeling methods, encompassing chemical probes-based labeling, metabolic labeling, and proximity-dependent labeling. We also provide a brief overview of their applications in the research of RNA biology. Finally, the perspectives of RNA labeling are also discussed.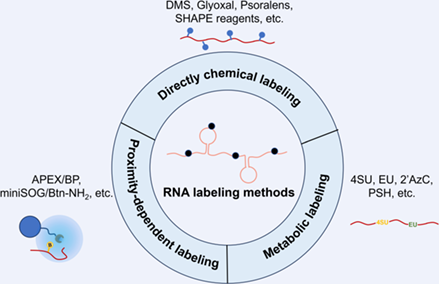
Key Scientists
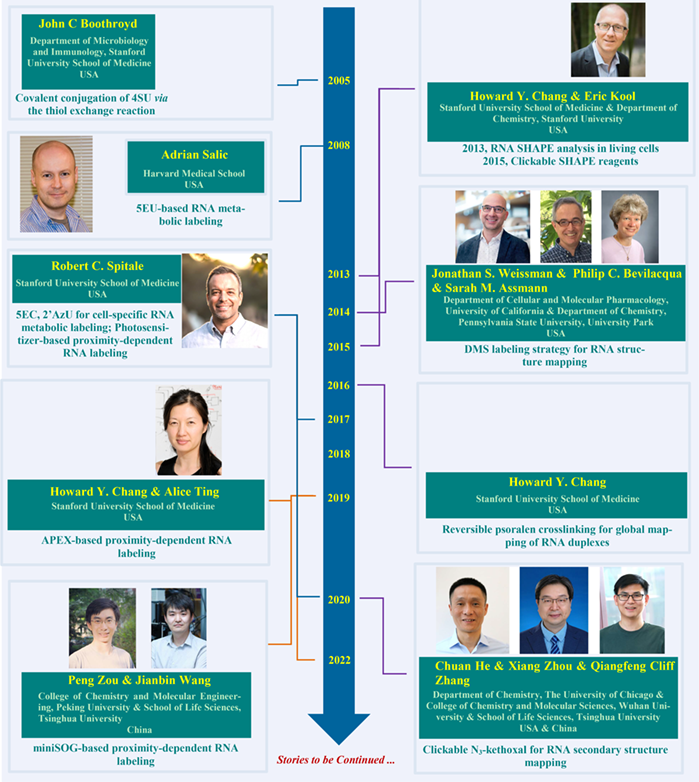
4SU was first used in1978 as a safer way to directly examine RNA synthesis. However, the enrichment of 4SU-labeled RNA seriously relied on mercurated cellulose columns, which are toxic. In 2005, Boothroyd et al. introduced a novel covalent-modification method of 4SU based on thiol exchange reaction. Then, Salic et al. applied 5EU, which is clickable, for RNA metabolic labeling in animals. Later, Spitale et al. designed 5EC and 2’AzU, which enabled cell specific RNA metabolic labeling under the help of specific enzyme. In 2013, directly chemical RNA labeling in living cell for RNA structure mapping was realized by Chang and Kool et al. using SHAPE reagent, which was further modified with an azide handle in 2015. Weissman, Bevilacqua and Assmann et al. introduced DMS probe that targets adenine and cytosine of RNA for RNA structure mapping in 2014. Then, Chang et al. exploited psoralen for RNA duplex mapping in 2016. Later, He, Zhou and Zhang et al. described another clickable probe N3-kethoxal targeting guanine for RNA secondary structure mapping. Besides, Chang and Ting et al. described a proximity-dependent RNA labeling method based on APEX in 2019, revealing subcellular RNA localization. Meanwhile, Zou and Wang et al. achieved proximity-dependent RNA labeling using miniSOG oxidase, which provided an alternative method to investigate the spatial transcriptome. Spitale et al. utilizing photosensitizer dibromofluorescein (DBF) to realize proximity-dependent RNA labeling and sequencing.
References
- 1 Morris, K. V.; Mattick, J. S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437.
- 2 Cech, T. R.; Steitz, J. A. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94.
- 3 Eddy, S. R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929.
- 4 Grosshans, H.; Filipowicz, W. Molecular biology - The expanding world of small RNAs. Nature 2008, 451, 414–416.
- 5 Carthew, R. W.; Sontheimer, E. J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655.
- 6 Mattick, J. S.; Amaral, P. P.; Carninci, P.; Carpenter, S.; Chang, H. Y.; Chen, L. L.; Chen, R. S.; Dean, C.; Dinger, M. E.; Fitzgerald, K. A.; Gingeras, T. R.; Guttman, M.; Hirose, T.; Huarte, M.; Johnson, R.; Kanduri, C.; Kapranov, P.; Lawrence, J. B.; Lee, J. T.; Mendell, J. T.; Mercer, T. R.; Moore, K. J.; Nakagawa, S.; Rinn, J. L.; Spector, D. L.; Ulitsky, I.; Wan, Y.; Wilusz, J. E.; Wu, M. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447.
- 7 Guerriertakada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease-P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857.
- 8 Pushpalatha, K. V.; Solyga, M.; Nakamura, A.; Besse, F. RNP components condense into repressive RNP granules in the aging brain. Nat. Commun. 2022, 13, 2782.
- 9 Shin, Y.; Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382.
- 10 Tanji, H.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Isobe, T.; Miyake, K.; Shimizu, T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015, 22, 109–115.
- 11 Langst, G.; Manelyte, L. Chromatin Remodelers: From Function to Dysfunction. Genes 2015, 6, 299–324.
- 12 Frankiw, L.; Baltimore, D.; Li, G. D. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 2019, 19, 675–687.
- 13 Karjee, S.; Mukherjee, S. K. RNAi suppressor: The hidden weapon of SARS-CoV. J. Biosci. 2020, 45, 99.
- 14 Li, L. X.; Luo, H. F.; Lim, D.; Han, L.; Li, Y.; Fu, X. D.; Qi, Y. J. Global profiling of RNA-chromatin interactions reveals co-regulatory gene expression networks in Arabidopsis. Nat. Plants 2021, 7, 1364–1378.
- 15 Lu, Z. P.; Zhang, Q. C.; Lee, B.; Flynn, R. A.; Smith, M. A.; Robinson, J. T.; Davidovich, C.; Gooding, A. R.; Goodrich, K. J.; Mattick, J. S.; Mesirov, J. P.; Cech, T. R.; Chang, H. Y. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279.
- 16 Aw, J. G. A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X. N.; Boon, K. L.; Tapsin, S.; Chan, Y. S.; Tan, C. P.; Sim, A. Y. L.; Zhang, T.; Susanto, T. T.; Fu, Z. Y.; Nagarajan, N.; Wan, Y. In vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617.
- 17 Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B. J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626.
- 18 Cai, Z. K.; Cao, C. C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z. C.; Hu, N. J.; Yu, X. H.; Chen, J.; Wang, L.; Yang, X. G.; He, S. M.; Xue, Y. C. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 2020, 582, 432–437.
- 19 Sun, L.; Xu, K.; Huang, W. Z.; Yang, Y. C. T.; Li, P.; Tang, L.; Xiong, T. L.; Zhang, Q. F. C. Predicting dynamic cellular protein-RNA interactions by deep learning using in vivo RNA structures. Cell Res. 2021, 31, 495–516.
- 20 Xie, J.; Zheng, J. F.; Hong, X.; Tong, X. X.; Liu, S. Y. PRIME-3D2D is a 3D2D model to predict binding sites of protein-RNA interaction. Commun. Biol. 2020, 3, 384.
- 21 Sun, L.; Fazal, F. M.; Li, P.; Broughton, J. P.; Lee, B.; Tang, L.; Huang, W. Z.; Kool, E. T.; Chang, H. Y.; Zhang, Q. F. C. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 2019, 26, 322–330.
- 22 Jones, C. P.; Ferre-D’Amare, A. R., Long-Range Interactions in Riboswitch Control of Gene Expression. Annu. Rev. Biophys. 2017, 46, 455–481.
- 23 Cheah, M. T.; Wachter, A.; Sudarsan, N.; Breaker, R. R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 2007, 447, 497–500.
- 24 Siwaszek, A.; Ukleja, M.; Dziembowski, A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014, 11, 1122–1136.
- 25 Savulescu, A. F.; Bouilhol, E.; Beaume, N.; Nikolski, M. Prediction of RNA subcellular localization: Learning from heterogeneous data sources. Iscience 2021, 24, 103298.
- 26 Nedelsky, N. B.; Taylor, J. P. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 272–286.
- 27 Autour, A.; Jeng, S. C. Y.; Cawte, A. D.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S. S. S.; Rueda, D.; Ryckelynck, M.; Unrau, P. J. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018, 9, 656.
- 28 Sato, S.-I.; Yatsuzuka, K.; Katsuda, Y.; Uesugi, M. Method for Imaging Live-Cell RNA Using an RNA Aptamer and a Fluorescent Probe. Methods Mol. Biol. 2018, 1649, 305–318.
- 29 Dean, K. M.; Palmer, A. E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 2014, 10, 512–523.
- 30 Caruthers, M. H. A brief review of DNA and RNA chemical synthesis. Biochem. Soc. Trans. 2011, 39, 575–580.
- 31 Rao, H.; Sawant, A. A.; Tanpure, A. A.; Srivatsan, S. G. Posttranscriptional chemical functionalization of azide-modified oligoribonucleotides by bioorthogonal click and Staudinger reactions. Chem. Commun. 2012, 48, 498–500.
- 32 Holstein, J. M.; Rentmeister, A. Current covalent modification methods for detecting RNA in fixed and living cells. Methods 2016, 98, 18–25.
- 33 Vaught, J. D.; Dewey, T.; Eaton, B. E. T7 RNA polymerase transcription with 5-position modified UTP derivatives. J. Am. Chem. Soc. 2004, 126, 11231–11237.
- 34 Li, M. Y. Optimization of N-hydroxysuccinimide ester coupling with aminoallyl-modified RNA for fluorescent labeling. Bioengineered 2020, 11, 599–606.
- 35 Liu, F.; Wang, H.; Li, S. H.; Bare, G. A. L.; Chen, X. M.; Wang, C.; Moses, J. E.; Wu, P.; Sharpless, K. B. Biocompatible SuFEx Click Chemistry: Thionyl Tetrafluoride (SOF4)-Derived Connective Hubs for Bioconjugation to DNA and Proteins. Angew. Chem. Int. Ed. 2019, 58, 8029–8033.
- 36 Sun, W.; Wang, N. X.; Liu, H. J.; Yu, B. C.; Jin, L.; Ren, X. J.; Shen, Y.; Wang, L. Genetically encoded chemical crosslinking of RNA in vivo. Nat. Chem. 2023, 15, 21–32.
- 37 Lempereur, L.; Nicoloso, M.; Riehl, N.; Ehresmann, C.; Ehresmann, B.; Bachellerie, J. P. Conformation of yeast 18s ribosomal-RNA-direct chemical probing of the 5’ domain in ribosomal-subunits and in deproteinized RNA by reverse-transcriptase mapping of dimethyl sulfate-accessible sites. Nucleic Acids Res. 1985, 13, 8339–8357.
- 38 Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2014, 505, 701–705.
- 39 Ding, Y. L.; Tang, Y.; Kwok, C. K.; Zhang, Y.; Bevilacqua, P. C.; Assmann, S. M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 2014, 505, 696–700.
- 40 Mitchell, D.; Ritchey, L. E.; Park, H.; Babitzke, P.; Assmann, S. M.; Bevilacqua, P. C. Glyoxals as in vivo RNA structural probes of guanine base-pairing. RNA 2018, 24, 114–124.
- 41 Noller, H. F.; Chaires, J. B. Functional modification of 16S ribosomal RNA by kethoxal. Proc. Natl. Acad. Sci. U. S. A. 1972, 69, 3115–3118.
- 42 Weng, X. C.; Gong, J.; Chen, Y.; Wu, T.; Wang, F.; Yang, S. X.; Yuan, Y. S.; Luo, G. Z.; Chen, K.; Hu, L. L.; Ma, H. H.; Wang, P. L.; Zhang, Q. F. C.; Zhou, X.; He, C. Keth-seq for transcriptome-wide RNA structure mapping. Nat. Chem. Biol. 2020, 16, 489–492.
- 43 Huang, J. G.; Zhao, R. Q.; Mo, J.; Wang, F.; Weng, X. C.; Zhou, X. N3-Kethoxal-Based Bioorthogonal Intracellular RNA Labeling. ChemBioChem 2021, 22, 1559–1562.
- 44 Knutson, S. D.; Sanford, A. A.; Swenson, C. S.; Korn, M. M.; Manuel, B. A.; Heemstra, J. M. Thermoreversible Control of Nucleic Acid Structure and Function with Glyoxal Caging. J. Am. Chem. Soc. 2020, 142, 17766–17781.
- 45 Zhao, R. Q.; Fang, X.; Mai, Z. B.; Chen, X.; Mo, J.; Lin, Y. Y.; Xiao, R.; Bao, X. C.; Weng, X. C.; Zhou, X. Transcriptome-wide identification of single-stranded RNA binding proteins. Chem. Sci. 2023, 14, 4038–4047.
- 46 Kuska, M. S.; Witham, A. A.; Sproviero, M.; Manderville, R. A.; Yazdi, M. M.; Sharma, P.; Wetmore, S. D. Structural Influence of C8-Phenoxy-Guanine in the NarI Recognition DNA Sequence. Chem. Res. Toxicol. 2013, 26, 1397–1408.
- 47 Xue, J. D.; Du, L. L.; Zhu, R. X.; Huang, J. Q.; Phillips, D. L. Direct Time-Resolved Spectroscopic Observation of Arylnitrenium Ion Reactions with Guanine-Containing DNA Oligomers. J. Org. Chem. 2014, 79, 3610–3614.
- 48 Voskresenska, V.; Wilson, R. M.; Panov, M.; Tarnovsky, A. N.; Krause, J. A.; Vyas, S.; Winter, A. H.; Hadad, C. M. Photoaffinity Labeling via Nitrenium Ion Chemistry: Protonation of the Nitrene Derived from 4-Amino-3-nitrophenyl Azide to Afford Reactive Nitrenium Ion Pairs. J. Am. Chem. Soc. 2009, 131, 11535–11547.
- 49 Feng, C.; Chan, D. L.; Joseph, J.; Muuronen, M.; Coldren, W.; Furche, F.; Hadad, C.; Spitale, R. Light-activated chemical probing of nucleobase solvent accessibility inside cells. Abstracts of Papers of the American Chemical Society 2018, 256, 276–283.
- 50 Kitamura, N.; Kohtani, S.; Nakagaki, R. Molecular aspects of furocoumarin reactions: Photophysics, photochemistry, photobiology, and structural analysis. J. Photochem. Photobiol. C-Photochem. Rev. 2005, 6, 168–185.
- 51 Kanne, D.; Straub, K.; Hearst, J. E.; Rapoport, H. Isolation and Characterization of Pyrimidine Psoralen Pyrimidine Photodiadducts from DNA. J. Am. Chem. Soc. 1982, 104, 6754–6764.
- 52 Zhang, M. J.; Li, K. P.; Bai, J. H.; Velema, W. A.; Yu, C. Q.; van Damme, R.; Lee, W. H.; Corpuz, M. L.; Chen, J. F.; Lu, Z. P. Optimized photochemistry enables efficient analysis of dynamic RNA structuromes and interactomes in genetic and infectious diseases. Nat. Commun. 2021, 12, 2344.
- 53 Wang, P. Y.; Sexton, A. N.; Culligan, W. J.; Simon, M. D. Carbodiimide reagents for the chemical probing of RNA structure in cells. RNA 2019, 25, 135–146.
- 54 Mitchell, D.; Renda, A. J.; Douds, C. A.; Babitzke, P.; Assmann, S. M.; Bevilacqua, P. C. In vivo RNA structural probing of uracil and guanine base-pairing by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC). RNA 2019, 25, 147–157.
- 55 Merino, E. J.; Wilkinson, K. A.; Coughlan, J. L.; Weeks, K. M. RNA structure analysis at single nucleotide resolution by selective 2 ’-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231.
- 56 Mortimer, S. A.; Weeks, K. M. Time-Resolved RNA SHAPE Chemistry. J. Am. Chem. Soc. 2008, 130, 16178–16180.
- 57 Steen, K. A.; Rice, G. M.; Weeks, K. M. Fingerprinting Noncanonical and Tertiary RNA Structures by Differential SHAPE Reactivity. J. Am. Chem. Soc. 2012, 134, 13160–13163.
- 58 Spitale, R. C.; Crisalli, P.; Flynn, R. A.; Torre, E. A.; Kool, E. T.; Chang, H. Y. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013, 9, 18–20.
- 59 Spitale, R. C.; Flynn, R. A.; Zhang, Q. C.; Crisalli, P.; Lee, B.; Jung, J. W.; Kuchelmeister, H. Y.; Batista, P. J.; Torre, E. A.; Kool, E. T.; Chang, H. Y. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486–490.
- 60 Kadina, A.; Kietrys, A. M.; Kool, E. T. RNA Cloaking by Reversible Acylation. Angew. Chem. Int. Ed. 2018, 57, 3059–3063.
- 61 Wang, S. R.; Wu, L. Y.; Huang, H. Y.; Xiong, W.; Liu, J.; Wei, L.; Yin, P.; Tian, T.; Zhou, X. Conditional control of RNA-guided nucleic acid cleavage and gene editing. Nat. Commun. 2020, 11, 91.
- 62 Xiao, L.; Jun, Y. W.; Kool, E. T. DNA Tiling Enables Precise Acylation-Based Labeling and Control of mRNA. Angew. Chem. Int. Ed. 2021, 60, 26798–26805.
- 63 Melvin, W. T.; Milne, H. B.; Slater, A. A.; Allen, H. J.; Keir, H. M. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur. J. Biochem. 1978, 92, 373–379.
- 64 Jao, C. Y.; Salic, A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 15779–15784.
- 65 Marguerat, S.; Lawler, K.; Brazma, A.; Bahler, J. Contributions of transcription and mRNA decay to gene expression dynamics of fission yeast in response to oxidative stress. RNA Biol. 2014, 11, 702–714.
- 66 Nainar, S.; Cuthbert, B. J.; Lim, N. M.; England, W. E.; Ke, K.; Sophal, K.; Quechol, R.; Mobley, D. L.; Goulding, C. W.; Spitale, R. C. An optimized chemical-genetic method for cell-specific metabolic labeling of RNA. Nat. Methods 2020, 17, 311–318.
- 67 Wang, D. Y.; Zhang, Y.; Kleiner, R. E. Cell- and Polymerase-Selective Metabolic Labeling of Cellular RNA with 2’-Azidocytidine. J. Am. Chem. Soc. 2020, 142, 14417–14421.
- 68 Hida, N.; Aboukilila, M. Y.; Burow, D. A.; Paul, R.; Greenberg, M. M.; Fazio, M.; Beasley, S.; Spitale, R. C.; Cleary, M. D. EC-tagging allows cell type-specific RNA analysis. Nucleic Acids Res. 2017, 45, e138.
- 69 Sawant, A. A.; Tanpure, A. A.; Mukherjee, P. P.; Athavale, S.; Kelkar, A.; Galande, S.; Srivatsan, S. G. A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA. Nucleic Acids Res. 2016, 44, e16.
- 70 Leibman, K. C.; Heidelberger, C. The metabolism of P32-labeled ribonucleotides in tissue slices and cell suspensions. J. Biol. Chem. 1955, 216, 823–830.
- 71 Boccaletto, P.; Machnicka, M. A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T. K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P. A.; Kotter, A.; Helm, M.; Bujnicki, J. M. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303-D307.
- 72 Mickute, M.; Nainyte, M.; Vasiliauskaite, L.; Plotnikova, A.; Masevicius, V.; Klimasauskas, S.; Vilkaitis, G. Animal Hen1 2’-O- methyltransferases as tools for 3’-terminal functionalization and labelling of single-stranded RNAs. Nucleic Acids Res. 2018, 46, e104.
- 73 Deen, J.; Vranken, C.; Leen, V.; Neely, R. K.; Janssen, K. P. F.; Hofkens, J. Methyltransferase-Directed Labeling of Biomolecules and its Applications. Angew. Chem. Int. Ed. 2017, 56, 5182–5200.
- 74 Burger, K.; Muhl, B.; Kellner, M.; Rohrmoser, M.; Gruber-Eber, A.; Windhager, L.; Friedel, C. C.; Dolken, L.; Eick, D. 4-Thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013, 10, 1623–1630.
- 75 Woodford, T. A.; Schlegel, R.; Pardee, A. B. Selective Isolation of Newly Synthesized Mammalian Messenger-Rna After in vivo Labeling with 4-Thiouridine or 6-Thioguanosine. Anal. Biochem. 1988, 171, 166–172.
- 76 Kenzelmann, M.; Maertens, S.; Hergenhahn, M.; Kueffer, S.; Hotz-Wagenblatt, A.; Li, L.; Wangt, S.; Ittrich, C.; Lemberger, T.; Arribas, R.; Jonnakuty, S.; Hollstein, M. C.; Schmid, W.; Gretz, N.; Grone, H. J.; Schutz, G. Microarray analysis of newly synthesized RNA in cells and animals. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 6164–6169.
- 77 Cleary, M. D.; Meiering, C. D.; Jan, E.; Guymon, R.; Boothroyd, J. C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 2005, 23, 232–237.
- 78 Michel, M.; Demel, C.; Zacher, B.; Schwalb, B.; Krebs, S.; Blum, H.; Gagneur, J.; Cramer, P. TT-seq captures enhancer landscapes immediately after T-cell stimulation. Mol. Syst. Biol. 2017, 13, 920.
- 79 Duffy, E. E.; Rutenberg-Schoenberg, M.; Stark, C. D.; Kitchen, R. R.; Gerstein, M. B.; Simon, M. D. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol. Cell 2015, 59, 858–866.
- 80 Herzog, V. A.; Reichholf, B.; Neumann, T.; Rescheneder, P.; Bhat, P.; Burkard, T. R.; Wlotzka, W.; von Haeseler, A.; Zuber, J.; Ameres, S. L. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 2017, 14, 1198–1204.
- 81 Schofield, J. A.; Duffy, E. E.; Kiefer, L.; Sullivan, M. C.; Simon, M. D. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods 2018, 15, 221–225.
- 82 Riml, C.; Amort, T.; Rieder, D.; Gasser, C.; Lusser, A.; Micura, R. Osmium-Mediated Transformation of 4-Thiouridine to Cytidine as Key to Study RNA Dynamics by Sequencing. Angew. Chem. Int. Ed. 2017, 56, 13479–13483.
- 83 Gasser, C.; Delazer, I.; Neuner, E.; Pascher, K.; Brillet, K.; Klotz, S.; Trixl, L.; Himmelstoss, M.; Ennifar, E.; Rieder, D.; Lusser, A.; Micura, R. Thioguanosine Conversion Enables mRNA-Lifetime Evaluation by RNA Sequencing Using Double Metabolic Labeling (TUC-seq DUAL). Angew. Chem. Int. Ed. 2020, 59, 6881–6886.
- 84 Baltz, A. G.; Munschauer, M.; Schwanhaeusser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; Wyler, E.; Bonneau, R.; Selbach, M.; Dieterich, C.; Landthaler, M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690.
- 85 Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B. M.; Strein, C.; Davey, N. E.; Humphreys, D. T.; Preiss, T.; Steinmetz, L. M.; Krijgsveld, J.; Hentze, M. W. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406.
- 86 Huang, R. B.; Han, M. T.; Meng, L. Y.; Chen, X. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E3879–E3887.
- 87 Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A. C.; Munschauer, M.; Ulrich, A.; Wardle, G. S.; Dewell, S.; Zavolan, M.; Tuschl, T. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 2010, 141, 129–141.
- 88 Schueler, M.; Munschauer, M.; Gregersen, L. H.; Finzel, A.; Loewer, A.; Chen, W.; Landthaler, M.; Dieterich, C. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol. 2014, 15, R15.
- 89 Bao, X. C.; Guo, X. P.; Yin, M. H.; Tariq, M.; Lai, Y. W.; Kanwal, S.; Zhou, J. J.; Li, N.; Lv, Y.; Pulido-Quetglas, C.; Wang, X. W.; Ji, L.; Khan, M. J.; Zhu, X. H.; Luo, Z. W.; Shao, C. W.; Lim, D. H.; Liu, X.; Li, N.; Wang, W.; He, M. H.; Liu, Y. L.; Ward, C.; Wang, T.; Zhang, G.; Wang, D. Y.; Yang, J. H.; Chen, Y. W.; Zhang, C. L.; Jauch, R.; Yang, Y. G.; Wang, Y. M.; Qin, B. M.; Anko, M. L.; Hutchins, A. P.; Sun, H.; Wang, H. T.; Fu, X. D.; Zhang, B. L.; Esteban, M. A. Capturing the interactome of newly transcribed RNA. Nat. Methods 2018, 15, 213–220.
- 90 Sletten, E. M.; Bertozzi, C. R. From Mechanism to Mouse: A Tale of Two Bioorthogonal Reactions. Acc. Chem. Res. 2011, 44, 666–676.
- 91 Nainar, S.; Beasley, S.; Fazio, M.; Kubota, M.; Dai, N.; Correa, I. R.; Spitale, R. C. Metabolic Incorporation of Azide Functionality into Cellular RNA. ChemBioChem 2016, 17, 2149–2152.
- 92 Van Rompay, A. R.; Norda, A.; Linden, K.; Johansson, M.; Karlsson, A. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 2001, 59, 1181–1186.
- 93 Tomkuviene, M.; Clouet-d’Orval, B.; Cerniauskas, I.; Weinhold, E.; Klimasauskas, S. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases. Nucleic Acids Res. 2012, 40, 6765–6773.
- 94 Hartstock, K.; Nilges, B. S.; Ovcharenko, A.; Cornelissen, N. V.; Pullen, N.; Lawrence-Dorner, A. M.; Leidel, S. A.; Rentmeister, A. Enzymatic or in vivo Installation of Propargyl Groups in Combination with Click Chemistry for the Enrichment and Detection of Methyltransferase Target Sites in RNA. Angew. Chem. Int. Ed. 2018, 57, 6342–6346.
- 95 Qin, W.; Cho, K. F.; Cavanagh, P. E.; Ting, A. Y. Deciphering molecular interactions by proximity labeling. Nat. Methods 2021, 18, 133–143.
- 96 Rhee, H. W.; Zou, P.; Udeshi, N. D.; Martell, J. D.; Mootha, V. K.; Carr, S. A.; Ting, A. Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339, 1328–1331.
- 97 Kim, D. I.; Jensen, S. C.; Noble, K. A.; Birendra, K. C.; Roux, K. H.; Motamedchaboki, K.; Roux, K. J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27, 1188–1196.
- 98 Branon, T. C.; Bosch, J. A.; Sanchez, A. D.; Udeshi, N. D.; Svinkina, T.; Carr, S. A.; Feldman, J. L.; Perrimon, N.; Ting, A. Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887.
- 99 Fazal, F. M.; Han, S.; Parker, K. R.; Kaewsapsak, P.; Xu, J.; Boettiger, A. N.; Chang, H. Y.; Ting, A. Y. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 2019, 178, 473–490.
- 100 Zhou, Y.; Wang, G.; Wang, P. C.; Li, Z. Y.; Yue, T. Q.; Wang, J. B.; Zou, P. Expanding APEX2 Substrates for Proximity-Dependent Labeling of Nucleic Acids and Proteins in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 11763–11767.
- 101 Huang, J. G.; Zhao, R. Q.; Qin, S. S.; Yang, S. X.; Li, W.; Mo, J.; Wang, F.; Du, Y. H.; Weng, X. C.; Zhou, X. 4-Thiouridine-Enhanced Peroxidase- Generated Biotinylation of RNA. ChemBioChem 2021, 22, 212–216.
- 102 Li, Y.; Aggarwal, M. B.; Nguyen, K.; Ke, K.; Spitale, R. C. Assaying RNA Localization in situ with Spatially Restricted Nucleobase Oxidation. ACS Chem. Biol. 2017, 12, 2709–2714.
- 103 Li, Y.; Aggarwal, M. B.; Ke, K.; Nguyen, K.; Spitale, R. C. Improved Analysis of RNA Localization by Spatially Restricted Oxidation of RNA-Protein Complexes. Biochemistry 2018, 57, 1577–1581.
- 104 Wang, P. C.; Tang, W.; Li, Z. Y.; Zou, Z. Y.; Zhou, Y.; Li, R.; Xiong, T. Y.; Wang, J. B.; Zou, P. Mapping spatial transcriptome with light- activated proximity-dependent RNA labeling. Nat. Chem. Biol. 2019, 15, 1110–1119.
- 105 Shu, X. K.; Lev-Ram, V.; Deerinck, T. J.; Qi, Y. C.; Ramko, E. B.; Davidson, M. W.; Jin, Y. S.; Ellisman, M. H.; Tsien, R. Y. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. PLoS Biol. 2011, 9, e1001041.
- 106 Engel, K. L.; Lo, H. Y. G.; Goering, R.; Li, Y.; Spitale, R. C.; Taliaferro, J. M. Analysis of subcellular transcriptomes by RNA proximity labeling with Halo-seq. Nucleic Acids Res. 2022, 50, e24.




