Chiral chromatography and surface chirality of carbon nanoparticles
[This article is part of the Special Issue: Chiral Materials.]
Funding information: University of Michigan; Momental Foundation; National Science Foundation, Grant/Award Number: DGE1841052
Abstract
Chiral carbon nanoparticles (CNPs) represent a rapidly evolving area of research for optical and biomedical technologies. Similar to small molecules, applications of CNPs as well as fundamental relationships between their optical activity and structural asymmetry would greatly benefit from their enantioselective separations by chromatography. However, this technique remains in its infancy for chiral carbon and other nanoparticles. The possibility of effective separations using high performance liquid chromatography (HPLC) with chiral stationary phases remains an open question whose answer can also shed light on the components of multiscale chirality of the nanoparticles. Herein, we report a detailed methodology of HPLC for successful separation of chiral CNPs and establish a path for its future optimization. A mobile phase of water/acetonitrile was able to achieve chiral separation of CNPs derived from L- and D-cysteine denoted as L-CNPs and D-CNPs. Molecular dynamics simulations show that the teicoplanin-based stationary phase has a higher affinity for L-CNPs than for D-CNPs, in agreement with experiments. The experimental and computational findings jointly indicate that chiral centers of chiral CNPs are present at their surface, which is essential for the multiple applications of these chiral nanostructures and equally essential for interactions with biomolecules and circularly polarized photons.
1 INTRODUCTION
Carbon nanoparticles (CNPs) have attracted extensive research interest for their unique physicochemical properties.1-3 CNPs with an average diameter of <10 nm share the desirable optical and electronic properties of typical semiconductor nanoparticles while avoiding disadvantages such as intrinsic toxicity from the use of heavy metals, harsh synthesis conditions, and expensive starting materials.4-6 CNPs and other nanocarbons display strong photoluminescence, easy functionalization, exceptional biocompatibility, high stability, and chemical inertness, making them useful for an array of applications including light emitting diodes,7 biosensors,8 photocatalysts,9 bioimaging,10 and drug delivery.11
Mirror asymmetric CNPs with multiple scales of chirality from molecular to nanoscale have the advantages of achiral CNPs with the added capability of chiral recognition for biochemical, biomedical, and sustainability applications involving biomolecues with similar hierarchy of chirality. One can expect that multiscale enantiomers of CNPs would elicit distinctly different, biological and pharmacological responses. For example, L-CNPs made from L-lysine remodeled the secondary structure of amyloid-β (aβ-42) peptides and inhibited key factors of pathogenesis, including cytolysis and amyloid fibril fibrillation, while D-CNPs made from D-lysine displayed little to no biological activity against the same peptide.12 Here we will refer to L- and D-NPs, when they are made from L- or D enantiomer of a molecular precursor, respectively. These notations represent only the chemical pathway for synthesis rather than the actual handedness at some scale. In another example, cysteine-derived L-CNPs upregulated glycolysis within human bladder cancer T24 cells, while D-CNPs did not display similar effects.13 Chiral CNPs synthesized from cysteine electrostatically interact with porphyrins to form supramolecular porphyrins–CNP hybrids, where the chirality of the CNPs transfers to the porphyrins and the CD signal are enhanced.14 Apart from graphene quantum dots,15 the localization of chiral centers and other chiral geometries within the CNPs remains an open question. CNPs were reported to have predominantly spherical geometries but could potentially have chiral geometries at multiple scales. Among those, CNPs are hypothesized to have the tetrahedral coordinated carbon centers inherited from chiral precursors but it is not clear whether these molecular structures can survive the high-temperature carbonization processes used for their preparation. Additionally, CNPs may also display nanoscale chirality associated both with ligand distribution at the interface and twist-distortions in the CNP interior.16 All of these structural features will result in a chiral environment around chromophores that will generate optical activity.17-19
The potential use of chiral CNPs for applications in biology, medicine, catalysis, and optics motivates us to determine whether the chiral centers in CNPs are located in the interior or at the interface of the particles. Importantly, the same question emerges when one needs to find reproducible methods for separating enantiomeric NPs. High performance liquid chromatography (HPLC) is the gold-standard method for chiral separations but has not been extended to resolving chiral nanoparticle solutions. Solution gradients are widely used for HPLC to separate analytes with disparate chemical composition. Chiral separations are typically conducted in isocratic conditions, making the mobile phase essential for separation yet difficult to optimize.20 Here, chiral resolution of cysteine-derived chiral CNPs was achieved using a simple water/acetonitrile mobile phase. To the best of our knowledge, this is the first report on enantioselective HPLC for the separation of enantiomeric NPs. The chiral stationary phase (CSP) employs the macrocyclic glycopeptide teicoplanin as the chiral selector grafted to silica gel via linkage chains and has been ideal for separating un-derivatized amino acids and other polar ionic chiral compounds.21-23 Molecular dynamics simulations show that D-CNPs preferentially interact with teicoplanin compared to L-CNPs, which is in agreement with the longer retention time of D-CNPs found experimentally. Chiral HPLC analysis of chiral nanostructures can determine enantiomeric nanoparticle purity and provide new methods for chiral separation and isolation. Simultaneously, these findings indicate that the surface of CNPs contains chiral centers.
2 EXPERIMENTAL SECTION
2.1 Materials
CNP precursors, namely, L-cysteine (L-cys) or D-cysteine (D-cys) and sodium hydroxide (NaOH), were purchased from Millipore Sigma. Racemic mixture of the amino acid cysteine (rac-cys) was prepared by mixing equal volumes of solution of L-cys and D-cys with a concentration of 1 mg/ml. Dialysis tubes with molecular weight cut off 1 kDa was obtained from Fischer Scientific. Acetonitrile (HPLC grade) was purchased from Millipore Sigma. All solutions were prepared using ultrapure water (18.2 MΩ·cm).
2.2 Synthesis of L- or D-CNPs
Chiral CNPs were synthesized asymmetrically by a hydrothermal method as previously described.24 L-cys or D-cys at 0.5 g and NaOH at 1 g were dissolved in 10 ml of water under ultrasonification for 20 min. The prepared solution was transferred into a 25 ml Teflon lined autoclave and heated at 120°C for 16 h. The mixture was dialyzed to remove unreacted material for 3 days. The final product was filtered against a 0.22 μm filter to remove large aggregates and then lyophilized for storage. Optically pure L- and D-CNPs were re-suspended to a final concentration of 1 mg/ml in ultrapure water prior to HPLC analysis.
2.3 Circular dichroism
CD spectra were acquired in a Jasco-815 spectrophotometer. The spectra were recorded from 190 to 500 nm, at 1 nm intervals, 1 nm bandwidth, and a scan speed of 100 nm/min.
2.4 Vibrational circular dichroism
VCD measurements were performed on CNP dispersions in heavy water (D2O) at a concentration of 33 mg/ml. A 100 μl drop was sandwiched between two BaF2 crystals separated by 50 μm Teflon spacer. MCT-V detector was used to acquire IR and VCD data in the range 1,800–850 cm−1 with a resolution of 4 cm−1 and a total of 200 and 1,000 accumulations, respectively. The sandwiched dispersion between BaF2 crystals was rotated along an axis coinciding with the direction of the beam at a constant speed to avoid settling of particles. Corresponding IR and VCD were plotted as absorbance (A) and differential absorbance of left and right circularly polarized light respectively with exclusion of 1,300 to 1,100 cm−1 range that corresponds to strong absorption from D2O.
2.5 Chiral HPLC analysis
Chiral CNPs were separated using an Agilent 6000 Series LC/MS instrument with an Astec CHIROBIOTIC T 10 cm × 4.6 mm (Millipore Sigma) chiral column. The mobile phase consisted of ultrapure water and acetonitrile (30:70 v/v) applied over 10 min at a flow rate of 1 ml min−1. UV detection was set to 205 nm.
2.6 Molecular dynamics
The affinity of L- and D-CNPs with teicoplanin was computed with biased all-atom simulations. The CHARMM force field, version 36, was employed, and the systems were solvated using explicit TIP3P water.25, 26 First, systems were minimized and equilibrated in the isothermal-isobaric ensemble for at least 150 ns, where a Langevin piston Nose-Hoover method (with a period of 200 and 50 fs decay) maintained a pressure of 100 kPa. A Langevin thermostat (with a characteristic time of 20 ps) was used to keep the temperature constant. A time step of 2 fs was employed to integrate the equations of motion, and hydrogen atoms are kept rigid via the SHAKE algorithm. Non-bonded short-range interactions smoothly approached 0 using an X-PLOR switching function between 1 and 1.2 nm, in conjunction with the particle mesh Ewald algorithm to evaluate long-range Coulombic forces. Nanoscale Molecular Dynamics (NAMD) software was used for running the simulations.27
Free energy surfaces were reconstructed using well-tempered Metadynamics simulations in the canonical ensemble over the course of 630 ns or more.28, 29 Gaussian-shaped bias (σ = 0.04 nm, initial height 0.2 kcal/mol) was deposited with pace 0.4 ps, and scaled using a bias factor of 10. Simulations were biased on the distance between teicoplanin's center of mass (COM) and the COM of each chiral CNP (dCOM). A harmonic wall was placed at 5 nm. The free energy was calculated as the average probability difference between the bound state (distance ≤ 2.5 nm) and the unbound state (distance ≥ 3 nm). PLUMED 2 was used for all simulation.30, 31 Visual Molecular Dynamics (VMD) was used for data visualization, and the MDAnalysis and Matplotlib Python libraries were used for plottin and data analysis.32-35
3 RESULTS AND DISCUSSION
Reversed-phase HPLC is the most common method for chiral analysis of amino acids and protein fragments. Analysis of protein fragments is achieved by chemical or enzymatic hydrolysis of proteins, followed by HPLC separation and, finally, analysis on mass spectrometry to identify protein sequence.36 Chiral CNPs are similar in size (2–10 nm) and molecular weight (103–105 g/mol) to proteins, yet if their chiral moieties are located on the surface, hydrolysis will be unnecessary for chiral analysis.37 Furthermore, biological molecules, namely, proteins, are known to adsorb onto the surface of CNPs changing the physicochemical properties of the NPs.38 Chiral HPLC analysis can be employed to characterize protein coronas by analyzing changes in retention time, which suggests changes in polarity or chirality in the case of CNP induced racemization. This makes chiral HPLC analysis a powerful tool to understand enantiomeric interactions between biological molecules and chiral CNPs.
Chiral CNPs were synthesized using a hydrothermal route using cysteine as both the carbon and chiral precursor,39 Figure 1A. Synthesis conditions (temperature and duration) were the same for synthesizing L-CNPs and D-CNPs to ensure their physicochemical properties were similar (size distribution, crystal structure, and surface functionalization). Subsequent characterization indicated that L-CNPs and D-CNPs have similar chemical structure and properties with exception of mirror symmetric relations between some chemical structures constituting the near-spherical particles. The main body of this manuscript will focus on the characterization of L-CNPs. More information on the physical and chemical properties of D-CNPs can be found in the supplemental information (SI).
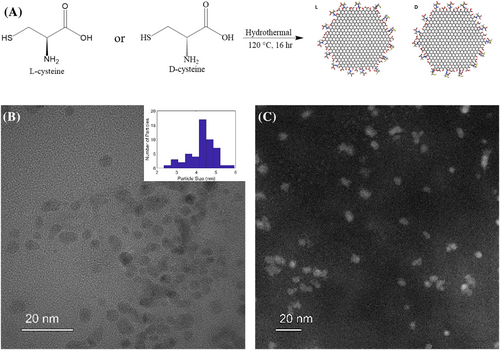
Morphological characteristics of the chiral CNPs were obtained using transmission electron microscopy (TEM) images that reveal both L- and D-CNPs have a size distribution between 2–7 nm (Figure 1B), which may be expected to be a challenge in their characterization and separation by chromatography. Scanning transmission electron microscopy high-angle annular dark field images (STEM-HAADF) images in Figure 1C confirm the size distribution and morphology of NPs as seen in TEM images.
The electrokinetic zeta-potential (ζ) of chiral CNPs was measured to determine the sign and estimate the magnitude of the averasurface charge on the particles (Figure S1). At pH 7, chiral CNPs display a strong negative charge of −27 ± 0.66 mV and −28.7 ± 0.26 mV for L-CNPs and D-CNPs, respectively. The negative charge is consistent with the presence of carboxyl groups at the edges.
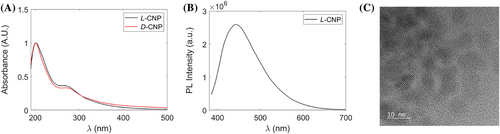
The optical properties of chiral CNPs were observed using UV–vis spectroscopy. For both L- and D-CNPs, two peaks were seen at 200 and 280 nm, which are attributed to the π-π* transition of the sp2 hybridized carbon network (Figure 2A).15 Fourier Transform Infrared Spectroscopy (FT-IR) was employed to determine the functional groups conjugated to the surface of the chiral CNPs (Figure 2B). The broad absorption band between 3,200 and 3,600 cm−1 are attributed to –OH and –NH stretching vibrations. The peak at 2,970 cm−1 indicates C-H bonds. The vibrational signals near 1,750 and 1,620 cm−1 were attributed to C=O and C=C, respectively.40 The presence of C-S bonds was seen by the absorption band at 1195 cm−1. FT-IR spectra confirmed the presence of carboxyl, amino, and thiol groups at the surface of the chiral CNPs.
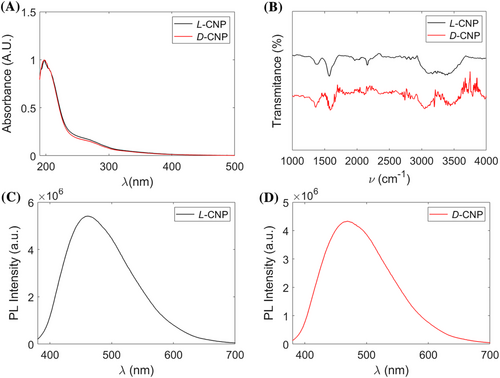
The photoluminescence spectra of chiral CNPs show the maximum emission wavelength between at ~475 nm after 365 nm excitation (Figure 2C,D). They indicate that the CNPs have typical light emission of conjugated aromatic compounds. The luminescence properties of these NPs are convenient for both analysis and biomedical applications.
Circular dichroism (CD) spectroscopy was used to investigate the chiroptical properties of the CNPs. CD spectra of L- and D- CNPs in Figure 3A reveal symmetrical and opposite high-energy peaks at 205 nm, which is the chiral signal from cysteine molecules. Both L- and D-CNPs gave rise to a new symmetrical CD signal at around 260 nm, which suggests there is an interaction between cysteine and the graphitic core of the CNPs.41
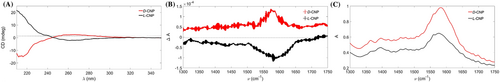
IR and vibrational circular dichroism (VCD) spectra were acquired to determine information about molcular scale chirality of CNPs. VCD data from previous reports of enantiomeric cysteine show mirror symmetry at the 1,621 cm−1 peak.42 Indeed, the mirror symmetric opposite band at 1,600 cm−1 in Figure 3B indicates that the chirality is transferred from the cysteine molecules to the CNPs. The IR spectrum in Figure 3C shows an antisymmetric stretching vibration at 1,600 cm−1 of carboxyl groups, which agrees which FT-IR data found in Figure 2C. Owing to the organic nature of CNPs the VCD spectra is on the order of 10−4.
The retention times of rac-cys and chiral CNPs can be found in Table 1. For rac-cys and chiral CNPs, the D- enantiomer had longer retention times compared to the L-enantiomer, which matches previous reports in literature.23 rac-cys contained two retention times, where L-cys resolved at 3.305 ± 0.145 min and D-cys shortly after at 3.368 ± 0.017 min. Figure 4A shows the corresponding chromatogram for rac-cys separation. The chiral CNPs eluted much faster than their cysteine precursors at 0.795 ± 0.046 min and 0.961 ± 0.022 min for L-and D- CNPs, respectively (Figure 4B). The longer retention times of the CNPs indicates their size prevents them interacting with chiral bonding sites deep within the CSP. This suggests that chiral ligands at the surface of the CNPs are the primary mechanism for enantioselective separation through polar ionic interactions.
| Analyte | Retention time 1 (min) | Retention time 2 (min) |
|---|---|---|
| rac-cys | 3.305 ± 0.145 | 3.368 ± 0.017 |
| L-CNP | 0.795 ± 0.046 | - |
| D-CNP | 0.961 ± 0.022 | - |
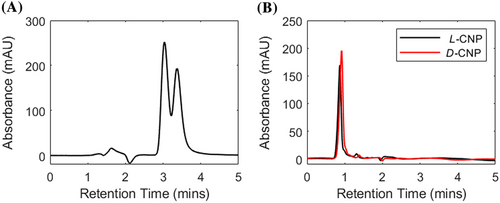
The peak resolution may be improved by adding organic or alcohol modifiers to the mobile phase. Reducing the polarity of the solvent will lower the elution strength and allow more π-π interactions between the chiral CNPs and the CSP. The size distribution of the chiral CNPs may also allow individual particles to have non-uniform interactions along the column. Further purification and fractionation of the CNPs can lower the size distribution and may improve enantioselective separation.
Chiral CNPs were captured after separation to ensure they maintain their morphology and physicochemical properties. UV–vis spectroscopy found in Figure 5A shows two similar peaks at 200 and 260 nm as seen in Figure 2 indicating minimal changes to the crystal structure of the CNPs. This point is confirmed by emission spectra of eluted CNPs. After 365 nm excitation, the same high-energy peak can be found between 450–500 nm similar to that in Figure 2. TEM microscopy of eluted CNPs maintain their size distribution of indicating the column separates by chirality and not by size. TEM also highlights the CNPs retain the lamellar structure of graphene, which proves CNPs are highly stable and resistant to hydrolysis. The stability of CNPs after separation allows the nanoparticles to be fractionated using HPLC for analysis or to be used in future experiments.
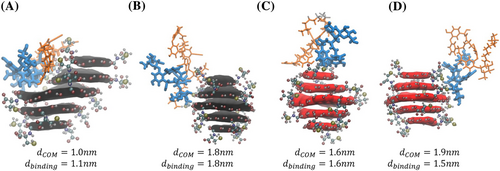
It has been established that teicoplanin is bound to aminopropyl silica gel through a bifunctional aliphatic isocyanate.43 Nonetheless, an incomplete understanding of the covalent grafting makes in silico investigation more difficult.44 Consequently, unbound teicoplanin was used to study its interaction with chiral CNPs using biased molecular dynamics (MD) simulations. Two simplified models were generated, one for teicoplanin with chiral NPs made from two cysteine enantiomers, under the mobile phase conditions stated above, to mimic the chromatographic conditions. Metadynamics simulations reveal a binding free energy for teicoplanin with L-and D-CNPs to be 0.35 ± 0.51 and −1.95 ± 0.64, respectively (Table 2), where the error is standard deviation. The higher binding energy for the teicoplanin/L-CNP system suggests a weaker or unfavorable interaction. Interestingly, L-CNPs become warped in close proximity to teicoplanin (Figure 6A); furthermore, the center of mass distance between teicoplanin's binding region and the L-CNP () tends to be larger than for the D-CNP when they are interacting. For example, teicoplanin's binding region orients itself closer to the D-CNP in Figure 6D, a possible rationalization for the difference in affinity (Table 2). Overall, MD simulations highlight the higher binding affinity between teicoplanin and D-CNPs, confirming the elution order found in experiments. This suggests MD simulations can be powerful tools for understanding the interactions of chiral NPs and chiral selectors.45, 46 Furthermore, MD simulations can be employed to screen multiple parameters to find suitable conditions for enhanced resolution between enantiomers.
| System | (kcal/mol) | Standard deviation |
|---|---|---|
| L-CNP | 0.35 | 0.51 |
| D-CNP | −1.95 | 0.64 |
4 CONCLUSIONS
Chiral CNPs made from two enantiomeric forms of cysteine were successfully separated using a water/acetonitrile mobile phase on an Astec CHIROBIOTIC T chiral column, which indicates the presence of chiral centers located on their surface. Besides the analytical importance enabling further development of biomedical and other applications of chiral nanoparticles, this finding is essential for the accurate physical picture of their interactions with cellular membranes47 and biomacromolecules.48 Various mirror asymmetric structures at multiple scales may also exist in the NP interior exemplified by the twist of their crystalline nanoparticle interior, helical conformation of molecular components, and a mirror-asymmetric surface. Unlike semiconductor and metal NPs,49, 50 the presence of these non-local components of multiscale chirality for CNPs remains uncertain. Furthermore, the condensation reactions leading to CNP formation are likely to lead to randomization and racemization of substituents around sp3 carbons. The ability of HPLC columns with teicoplanin stationary phase to separate these L- and D-CNPs give strong indication that L- and D-centers of cysteine are retained. Additionally, these data indicate that chiral CNPs are hydrolytically stable and resist degradation into their precursor cysteine. The dominat forces behind chiral separations were steric hindrance and polar ionic. MD simulations were employed to investigate the interaction between chiral NPs and the chiral selector. This technique provides a new method for detecting and quantifying chiral NPs. While providing foundation for future chiral HPLC analysis of chiral NPs, chiral HPLC analysis of NPs can also provide an analytical toolbox for future studies of chiral recognition mechanisms and methods for interacting with different chiral centers.
ACKNOWLEDGMENTS
This work was funded by the National Science Foundation Graduate Research Fellowship Program Grant No. DGE1841052. Parts of this work were also supported by the Momental Foundation through the Mistletoe Research Fellowship program. The authors also acknowledge the University of Michigan, College of Engineering, for financial support of the BlueSky Initiative, Michigan Center for Materials Characterization for use of the instruments, and Dr. Tao Ma for acquiring the TEM images. Thank you to Dr. Emine Sumeyra Turali Emre for generating achiral CNP models. The graphical abstract was created with BioRender.com.
CONFLICT OF INTEREST
The authors declare they have no conflict of interest related to this work.
Open Research
DATA AVAILABILITY STATEMENT
Data are available upon request from authors.




