A novel mutation of SETBP1 in atypical chronic myeloid leukemia transformed from acute myelomonocytic leukemia
Key Clinical Message
To investigate an oncogenic mutation of SETBP1 in the evolution from acute myelomonocytic leukemia (M4) to secondary aCML. Clinical data and molecular studies were analyzed of paired aCML and 'normal'DNA from a case with M4. We identified a mutation in SETBP1 (encoding a p.Asp868Ala alteration). The analysis of paired sample indicated that SETBP1 mutation was acquired during leukemic evolution.
Introduction
aCML is a rare myeloproliferative neoplasm with a combination of myeloproliferation and dysplastic hematopoiesis in clinical manifestation. The pathogenetic basis of aCML has remained elusive. Some recent studies have increased our understanding of the genetic drivers of MPN 1-7, More recently, substantial progress has been discovered in the understanding of the pathogenic gene mutations driving aCML, such as SETBP1 8-13, CSF3R 1-3. aCML occurring as a secondary malignancy is rare. Recently, we identified an oncogenic mutation in the SETBP1 in aCML. Here, we report a rare case of aCML in a patient after chemotherapy for M4. Herein, we discussed the potential impact of the finding on the pathogenic gene of these disorders.
Case History/Examination
A 63-year-old man was referred to our hospital because of increases in blood cell counts. Laboratory tests showed a hemoglobin level of 7.2 g/dL, leukocytes 28 × 109/L, and platelets 76 × 109/L. There was no splenomegaly, hepatomegaly, lymphadenopathy, or obvious morphological abnormalities of hemocyte (include bone marrow and peripheral blood) were found. Examination of the bone marrow shows a significant myeloid hyperplasia with full myeloblasts (35% in the peripheral blood), POX (Strongly positive), NSE (100% positive), and restrain by NaF. Chromosome analysis with G-banded karyotype of the unstimulated bone marrow cells showed 46, XY, in all 24 metaphase spreads. The patient was diagnosed with acute myelocytic leukemia (AML-M4, Fig. 1). The patient was treated with CAG (ACR, Ara-c, G-CSF, more than 60 years of age and low doses of chemotherapy regimens) chemotherapy regimens, at this dose, a hematological complete remission (CR) after each chemotherapy was attained, after 6× CAG chemotherapy regimens, after the last stage of chemotherapy (remain complete remission, WBC count was normal), separately. About 3 months later, peripheral blood count revealed a workup for leukocytosis (16.4 × 109/L) and thrombocytosis (378 × 109/L). Examination of peripheral blood smear revealed blast cells (3.5%), immature granulocytes (16%), and the phenomenon of dysplasia in granulopoiesis. Neither monocytosis (2%), basophilia (1%), nor eosinophilia (0.5%) was seen. The neutrophil alkaline phosphatase (NAP) score was 3 (control, 78). Bone marrow biopsy was observed with an obvious myeloid proliferation (include mature neutrophils and neutrophil precursors), granulocytic dysplasia, and marked megakaryocytosis (include some small megakaryocytes) (Fig. 1). Genetic mutation analysis of the bone marrow cells showed as follows (Table 1). No structural or numerical abnormalities of chromosome were detected at the stage of aCML. Other clinical features include splenomegaly and anemia. The patient was diagnosed with aCML. Limited treatment with hydroxyurea achieved a partial hematologic response. The case of aCML and 43 additional cases of AML (M4) were confirmed to meet WHO-2008 14 diagnostic criteria. No SETBP1 mutation was found in any of the AML. 2/43 AML (M4) cases exhibited WT-1 mutations (Table 2). The present study was adhered to the tenets of the Declaration of Helsinki.
| Gene | M4 | aCML |
|---|---|---|
| AML1/ETO | (+) | (−) |
| JAK2-V617F | (−) | (−) |
| BCR/ABL (P190/210) | (−) | (−) |
| WT-1 | (+) | (+) |
| MPL W515L/K | (−) | (−) |
| FIP1L1-PDGFRα | (−) | (−) |
| ETV6-PDGFRβ | (−) | (−) |
| SETBP1 | (−)a | (+) |
| CSF3R | (−) | (−) |
- (−): no mutation was detected; (+): the presence of a mutation.
- a Mutations detected after each CAG chemotherapy on remission stage.
| Gene | Number of mutated samples (n = 43) |
|---|---|
| SETBP1 | 0/43 |
| WT-1 | 2/43 |
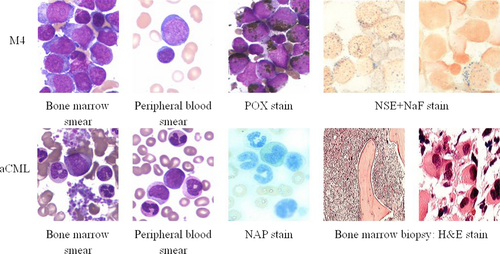
A Novel Mutation of SETBP1 Gene in aCML
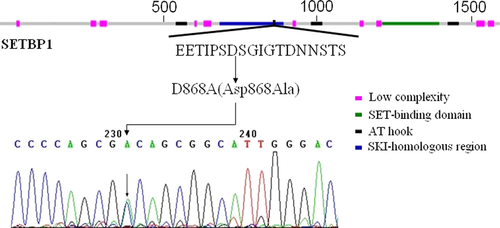
SETBP1 mutation of paired aCML (bone marrow) and ‘normal' DNA from a case with M4 was detected by Sanger sequencing. SETBP1 mutation (showed a heterozygous mutation A/C) was located in the part of the gene encoding the SKI homologous (aa 706–917) domain (Fig. 2). SETBP1-mutant showed significantly higher SETBP1 mRNA expression levels compared to wild-type case without SETBP1 overexpression.
Discussion
Recently, substantial progress has been found for the understanding of the pathogenic gene mutations driving aCML, such as CBL 5-7, RAS 8, PDGFRB 4, SETBP1 8-13, TET2 5-7, and CSF3R 1-3. So far, SETBP1 has been identified the most specific genetic mutation in the aCML 8, 9. In an AML1/ETO-positive patient, a potential mechanism regulating the evolution from M4 to secondary aCML with mutation of SETBP1 will be discussed.
Insights into the evolution of a case from M4 to aCML, clinical samples available to successfully analyze were collected serially at different stages of clinical time points. We performed exon sequencing and identified a new mutation in SETBP1 (encoding a p.Asp868Ala alteration), which was subsequently confirmed by Sanger sequencing. No SETBP1 mutation was found in 43 AML samples, indicated that there was a low frequency of SETBP1 mutation in AML. Furthermore, previous studies demonstrated that SETBP1 is mutated at lower frequencies in other MDS/MPN 9.
The same and closely neighboring SETBP1 mutations encoding changes in have been reported, such as E858K in aCML 8, 9, D868A in SGMFS 10, D868G/Y 9, 11, and S869R 9 in myeloid malignancies, D868N in SGMFS 10, aCML 8, 9, 12, JMML 13 and MDS 12, S869N in MDS 12, and myeloid malignancies 11, all mutations clustered in the SKI homologous region of SETBP1 are within a stretch of 22 aa (aa 858–880). But so far no SETBP1 (D868A) mutation has been identified in aCML patient. SETBP1 interacts with SET and decreases PP2A activity 9, 15-17. When phosphorylated, the SKI homologous region will be bound by the β-TrCP1 and thus leads to the degradation 8. However, the SETBP1 mutants disrupt this progression and lead to the increased SETBP1 and SET expression, which lower PP2A activity and show significantly more efficient colony formation and faster proliferation of cells 8. In addition, previous studies have confirmed that mutant SETBP1 immortalized myeloid leukemia cell 18, our finding is consistent with the gain of leukemogenic function due to SETBP1 mutation. In agreement with these findings, the p.Asp868Ala alteration abrogated a site for degradation, and cells expressing this mutant exhibited higher amounts of SETBP1 mRNA relative to the expressing the AML-M4 CR SETBP1 mRNA. We also propose that mutation could increase protein stability, resulting in greater protein amounts (similar to overexpression of SETBP1 mRNA) 8. These cause the gain of function associated with myeloid leukemic transformation and may be relevant to the pathogenesis of aCML.
We have confirmed the paired samples of the other genetic variation including WT-1. These observations indicated an association of WT-1 mutated 19 with SETBP1 mutated cases in aCML patients. This signifies a potential influence of the other oncogene genes on pathogenesis of aCML. Whereas JAK2V617F-negative MPN may be observed with MPLW515 20, 21, FIP1L1-PDGFRα 22, ETV6-PDGFRβ 4 mutations, and they are all negative in our sample.
None of the M4 had SETBP1 mutation at the time of initial stage, clearly revealed that SETBP1 mutation was acquired with the leukemic evolution. We propose that SETBP1 may be a high risk for leukemic evolution. Additional screening for SETBP1 mutations might be helpful for the diagnosis of aCML, even though this particular observation requires further confirmation in a larger cohort of WHO-defined aCML.
Otherwise, what indicates that aCML does not precede the AML occurrence? We postulate that the SETBP1 mutation had arisen after induction chemotherapy at AML CR. Though equally likely, the patient might have had undiagnosed SETBP1 causing aCML that was transiently out-competed by a RUNXI-RUNX1T1 AML clone. At CR, chemotherapy eradicated the AML1-ETO clone, leaving the SETBP1 clone detectable again. There are now many examples of AML that develop on a background of myeloproliferative neoplasm – for example, JAK2 V617F mutation – without detection of the mutation in AML blast cells, suggesting that the AML develops from a distinct stem cell or progenitor, possibly on a background of other mutations (such as TET2 mutation) 23, 24.
In conclusion, we identified a new mutation in SETBP1 (encoding a p.Asp868Ala alteration) of the aCML, SETBP1 mutation may be acquired during leukemic evolution.
Acknowledgments
We gratefully acknowledge the patient for their participation in our study. We thank Dr. Xiao Bo Nie for the critical reading of this manuscript.
Conflict of Interest
The authors declare that the paper is the authors' original work, has not been submitted to or published by another journal, and there are no competing financial interests or conflicts of interest in association with this research.




