Inhibiting H3K27 Demethylases Downregulates CREB-CREBBP, Overcoming Resistance in Relapsed Acute Lymphoblastic Leukemia
Funding: This work was supported by Agència de Gestió d'Ajuts Universitaris i de Recerca-Generalitat de Catalunya, 2021-SGR-00284 (C.A and J.M). Ministerio de Ciencia e Innovación, Proy. de Generación del Conocimiento 2021, PID2021-128644OB-I0 (C.A and J.M). Deutsche Carreras Leukämie Stiftung, DJCLS 01R/2016 (L.B. and C.D.B.), DJCLS 19R/2022 (J.L, C.A., J.M. and C.E.).
ABSTRACT
Background
CREB binding protein (CREBBP) is a key epigenetic regulator, altered in a fifth of relapsed cases of acute lymphoblastic leukemia (ALL). Selectively targeting epigenetic signaling may be an effective novel therapeutic approach to overcome drug resistance. Anti-tumor effects have previously been demonstrated for GSK-J4, a selective H3K27 histone demethylase inhibitor, in several animal models of cancers.
Methods
To characterize the effect of GSK-J4, drug response profiling, CRISPR-Dropout Screening, BH3 profiling and immunoblotting were carried out in ALL cell lines or patient derived samples.
Results
Here we provide evidence that GSK-J4 downregulates cyclic AMP-responsive element-binding protein (CREB) and CREBBP in B-cell precursor-ALL cell lines and patient samples. High CREBBP expression in BCP-ALL cell lines correlated with high GSK-J4 sensitivity and low dexamethasone sensitivity. GSK-J4 treatment also induced Bcl-2 and Bcl-XL dependency and apoptosis.
Conclusions
This study proposes H3K27 demethylase inhibition as a potential treatment strategy for patients with treatment-resistant ALL, using CREBBP as a biomarker for drug response and combining GSK-J4 with venetoclax and navitoclax as synergistic partners.
1 Introduction
Implementing contemporary treatment concepts has raised relapse-free survival to > 80% in children with acute lymphoblastic leukemia (ALL) [1]. Despite significant progress in risk-adapted treatment concepts, the emergence of drug resistance at relapse remains a major challenge to the successful treatment of ALL. At relapse, pediatric B-cell precursor (BCP)-ALL is enriched with somatic mutations in the genes for several epigenetic regulators, including the CREB binding protein (CREBBP), lysine demethylase 6A (KDM6A, previously known as UTX), lysine methyltransferase 2D (KMT2D) and SET domain containing 2, histone lysine methyltransferase (SETD2), suggesting the role of epigenetic regulators in development of treatment resistance [2, 3]. CREBBP alterations consisting of either sequence mutations or deletions occur in 18.3% of patients with relapsed ALL [2]. Dysregulation of the cAMP-response element binding protein 1 (CREB1), is implicated in ALL drug resistance (e.g., to glucocorticoids) [4]. CREB1 is a transcription factor involved in epigenetic regulation of key epigenetic elements (e.g., enhancer of zeste 2 polycomb repressive complex 2 subunit; EZH2), and its overexpression leads to increased histone 3 lysine 27 trimethylation (H3K27me3) [5]. CREBBP forms complexes with CREB1 or E1A-binding protein (EP300), whose histone acetyltransferase activity modulates cellular processes such as growth, survival, differentiation, and hematopoiesis, by regulating transcription factors such as MYC [6] and glucocorticoid responsive genes [4]. These findings suggest that targeting key epigenetic regulators in ALL [7] could be a new approach to for the treatment of relapsed ALL patients, and raises the need to investigate novel inhibitors in preclinical settings capable of selectively targeting epigenetic regulators that can overcome drug resistance.
2 Material and Methods
Detailed information on applied material and methods is provided in Appendix S1.
3 Results
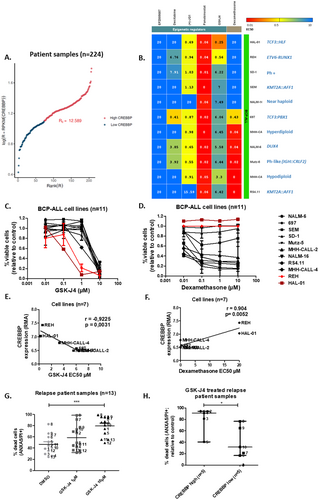
RNAseq analysis in a cohort of 224 pediatric patients with relapsed BCP-ALL identified heterogeneous CREBBP expression, with 19 relapses (8.5% of cases) overexpressing CREBBP (Figure 1A). CREBBP RNA levels correlated positively with EP300 levels and negatively with CREB1 (encoding CREB protein; Figure 1A, Figure S1) levels. To identify novel targetable vulnerabilities regulating the relapse ALL epigenome, a drug screen including 5 epigenetic regulators and dexamethasone was carried out in BCP-ALL cell lines (n = 11, Figure 1B). Among the most compelling drug sensitivity patterns identified, was that the dexamethasone-resistant BCP-ALL cell lines, HAL-01, and REH, (half-maximal effective concentrations; EC50 > 20 μM) were the most sensitive to GSK-J4 (EC50 = 0.25 μM and 0.56 μM, median EC50 = 6.2 μM, Figure 1C,D). GSK-J4 selectively inhibits KDM6A and KDM6B H3K27 demethylases. GSK-J4 EC50 values were significantly inversely correlated with CREBBP RNA expression in BCP-ALL cell lines (r = −0.92, p = 0.003, Figure 1E). Dexamethasone EC50 values were significantly correlated with higher CREBBP RNA expression (r = 0.9, p = 0.005, Figure 1F), suggesting that cell lines expressing higher levels of CREBBP are less sensitive to dexamethasone but more sensitive to GSK-J4. Treatment with GSK-J4 reduced BCP-ALL cells (n = 4 lines) at the cell cycle phase S/G2 transition and, although not significantly, enhanced the proportion of cells in G1, suggesting that GSK-J4 treatment arrests the cell cycle at G1 (Figure S2). To better reflect biological and clinical heterogeneity in BCP-ALL cases, patient-derived samples (n = 13, patient information in Table S1), obtained after first or second relapse, were treated in vitro with GSK-J4. GSK-J4 significantly induced apoptosis (p = 0.0005) in patient-derived BCP-ALL cells (Annexin V and PI staining, Figure 1G). In accordance with our findings in cell lines, sensitivity to GSK-J4 was significantly higher (59.6% median increase, p = 0.03) in patient-derived samples expressing higher CREBBP RNA levels compared to patient samples with lower CREBBP levels (Figure 1H). Overall, our data suggest high CREBBP expression is a biomarker determining dexamethasone-resistant BCP-ALL cell sensitivity to GSK-J4.
To study the interaction between GSK-J4 treatment and the CREB1/CREBBP complex, western blot analysis was carried out in different BCP-ALL cell lines (n = 4) and 2 patient-derived samples. GSK-J4 treatment downregulated CREB1 and KDM6A protein levels across the different ALL cells tested (Figure 2A, Figure S3). Except for HAL-01, which is not detected by the CREBBP antibody because it harbors a CREBBP mutation (c.6299 T > G), all the BCP-ALL cells demonstrated downregulated CREBBP protein levels in response to GSK-J4 treatment (Figure 2A, Figure S3). To better understand the molecular mechanisms underlying GSK-J4 sensitivity, we conducted a CRISPR drop-out screen [8] in 697, NALM-6, and REH cell lines expressing Cas9. A comparison between wild-type and knockout cells revealed a dependency on EP300 and CREBBP across all cell lines, as well as on BCL2 (negative beta values; Figure 2B). Dependence to CREBBP was seen highest in GSK-J4 sensitive cell line REH, compared to less sensitive cells NALM-6 and 697. In GSK-J4-treated cells, EP300 exhibited a protective effect (positive beta values, Figure 2C). Notably, in REH cells, which are sensitive to GSK-J4, synthetic lethality was observed in GSK-J4 treated cells harboring CREBBP KO and BCL2 KO. These findings further underscore the role of CREBBP in mediating GSK-J4 sensitivity, identifying it as a critical signaling element, essential for ALL cell survival.
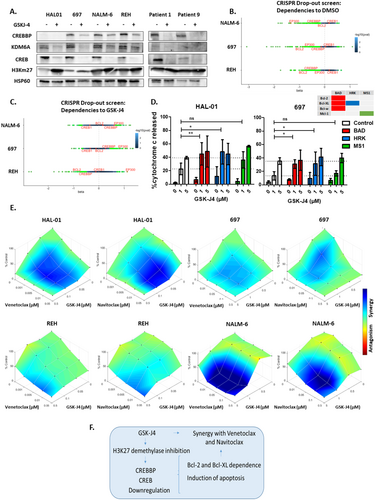
Dynamic BH3 profiling [9] is a functional assay allowing the study of underlying mechanisms of cellular adaptations to overcome apoptosis in different ALL cells before and after treatment [10]. We performed dynamic BH3 profiling in HAL01 and 697 cell lines treated with GSK-J4 together with either BAD, HRK or MS1 peptides that respectively target the Bcl-2/Bcl-XL/Bcl-w, Bcl-XL only or Mcl-1 anti-apoptotic proteins. Treatment with BAD and HRK peptides but not MS1, significantly increased cytochrome C release after treatment with 1 μM GSK-J4 in both cell lines (Figure 2D), indicating the cells are dependent on Bcl-2 and BCL-XL to resist apoptosis induced by GSK-J4. This suggests a potential targetable vulnerability that could be overcome by combining GSK-J4 with an appropriate BH3 mimetic drug. Consequently, we analyzed synergy between GSK-J4 combined with either venetoclax (Bcl-2 inhibitor) or navitoclax (Bcl-2/Bcl-XL inhibitor). Serial dilutions of these BH3 mimetics, 20 combined concentrations of venetoclax/navitoclax alone (n = 4), GSK-J4 alone (n = 3) and all combinations of these, were applied to BCP-ALL cell lines (n = 4). Using the HSA reference model, synergy was analyzed in 4 BCP-ALL cell lines, identifying at least one significant synergistic combination in each compound and cell line tested (Figure 2E, Table S2). The synergy between BH3 mimetics and GSK-J4 is strongest in the NALM-6 cell line, which is least sensitive to GSK-J4 alone, and weaker in the most sensitive cell line, REH (8 and 1–2 significant synergistic combinations respectively). Our results demonstrate that inhibiting H3K27me3 demethylases induces a Bcl-2 and Bcl-XL dependency in cell lines, both sensitive and resistant to GSK-J4, leading to a synergistic interaction between GSK-J4 and the BH3 mimetics, venetoclax, and navitoclax.
4 Discussion
Previous studies have reported the anti-tumor effect of GSK-J4 in xenograft mouse models of acute myeloid leukemia (AML) [11] and breast cancer [12], and for GSK-J4 in combination with oxaliplatin in xenograft mouse models of colorectal cancer [13]. Higher KDM6B expression levels was linked to worse survival in diffuse large B-cell lymphoma patients, and cell lines with mutations in CREBBP/EP300 presented an increased sensitivity to GSK-J4 treatment [14]. Here we propose a model in which GSK-J4 treatment downregulates CREB and CREBBP expression in ALL cells, which subsequently induces cell cycle arrest and even apoptosis, with higher sensitivity seen in cells expressing high CREBBP levels. High CREBBP expression was also correlated with heightened GSK-J4 sensitivity and diminished dexamethasone sensitivity in multiple ALL cell lines on RNA and protein levels in the recently published FORALL [15] cell line database (Figure S4). Our data is in accordance with previous results from AML cell lines, where CREB1 protein levels decreased upon GSK-J4 treatment, a process regulated by CREB1 phosphorylation at the Ser133 residue by protein kinase A (PKA) and leading to its proteasomal degradation [16].
We provide evidence here that inhibiting H3K27me3 demethylases is a potential treatment strategy for relapse patients with ALL who do not respond to dexamethasone. We propose a strategy (Figure 2F) in which CREBBP expression can be used as a potential biomarker to indicate response to GSK-J4, which induces a Bcl-2 and Bcl-XL dependency that can be targeted by combining GSK-J4 with clinically available [17, 18] synergistic partners venetoclax and navitoclax.
Author Contributions
Juan Lazaro-Navarro: conceptualization (lead), data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), writing – original draft (lead), writing – review and editing (lead). Clara Alcon: data curation (equal), formal analysis (equal), methodology (equal), writing – review and editing (equal). Mathurin Dorel: methodology (equal), writing – review and editing (equal). Lina Alasfar: data curation (equal), formal analysis (equal), writing – review and editing (equal). Lorenz Bastian: conceptualization (equal), funding acquisition (equal), supervision (equal). Claudia Baldus: funding acquisition (equal), supervision (equal), writing – review and editing (equal). Kathy Astrahantseff: writing – original draft (equal), writing – review and editing (lead). Marie-Laure Yaspo: supervision (equal), writing – review and editing (equal). Joan Montero: funding acquisition (equal), methodology (equal), supervision (equal), writing – review and editing (equal). Cornelia Eckert: conceptualization (equal), funding acquisition (lead), project administration (lead), supervision (lead), writing – original draft (equal), writing – review and editing (equal).
Acknowledgements
We to acknowledge Mila Valli for performing cloning of plasmids.
Ethics Statement
All patients were enrolled in the IntReALL SR/HR 2010 trials or the ALL-REZ BFM registry, approved by the State Office for Health and Social Affiars Berlin and the Institutional Review Board of the Charité Universitätsmedizin Berlin, Berlin, Germany respectively (ClinicalTrials.gov identifier: NCT01802814/NCT03590171). Written informed consent was obtained from patients or guardians.
Conflicts of Interest
J.M. is co-inventor of dynamic BH3 profiling (patented by Dana-Faber Cancer Institute) and has received royalties, was a paid consultant for Oncoheroes Biosciences and Vivid Biosciences, is an unpaid board member for The Society for Functional Precision Medicine, and he is currently collaborating with AstraZeneca. No potential conflicts of interest were disclosed by the other authors.
Open Research
Data Availability Statement
The data underlying this study will be made available upon request to the authors via email.