Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma
Chang Zhang, Qi Sun and Xu Zhang contributed equally to this work.
Abstract
Background
Epigenetic alterations have been shown to contribute immensely to human carcinogenesis. Dynamic and reversible N6-methyladenosine (m6A) RNA modification regulates gene expression and cell fate. However, the reasons for activation of KIAA1429 (also known as VIRMA, an RNA methyltransferase) and its underlying mechanism in lung adenocarcinoma (LUAD) remain largely unexplored. In this study, we aimed to clarify the oncogenic role of KIAA1429 in the tumorigenesis of LUAD.
Methods
Whole-genome sequencing and transcriptome sequencing of LUAD data were used to analyze the gene amplification of RNA methyltransferase. The in vitro and in vivo functions of KIAA1429 were investigated. Transcriptome sequencing, methylated RNA immunoprecipitation sequencing (MeRIP-seq), m6A dot blot assays and RNA immunoprecipitation (RIP) were performed to confirm the modified gene mediated by KIAA1429. RNA stability assays were used to detect the half-life of the target gene.
Results
Copy number amplification drove higher expression of KIAA1429 in LUAD, which was correlated with poor overall survival. Manipulating the expression of KIAA1429 could regulate the proliferation and metastasis of LUAD. Mechanistically, the target genes of KIAA1429-mediated m6A modification were confirmed by transcriptome sequencing and MeRIP-seq assays. We also revealed that KIAA1429 could regulate BTG2 expression in an m6A-dependent manner. Knockdown of KIAA1429 significantly decreased the m6A levels of BTG2 mRNA, leading to enhanced YTH m6A RNA binding protein 2 (YTHDF2, the m6A “reader”)-dependent BTG2 mRNA stability and promoted the expression of BTG2; thus, participating in the tumorigenesis of LUAD.
Conclusions
Our data revealed the activation mechanism and important role of KIAA1429 in LUAD tumorigenesis, which may provide a novel view on the targeted molecular therapy of LUAD.
Abbreviations
-
- 3’UTR
-
- 3’untranslated region
-
- ANOVA
-
- Analysis of Variance
-
- BSA
-
- bovine serum albumin
-
- cDNA
-
- complementary DNA
-
- CNVs
-
- copy number variations
-
- DAPI
-
- 4’,6-diamidino-2-phenylindole
-
- DAVID
-
- Database for Annotation, Visualization, and Integrated Discovery
-
- DEGs
-
- differentially expressed gene
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- EDTA
-
- Ethylene Diamine Tetraacetic Acid
-
- EdU
-
- 5-ethynyl-2’-deoxyuridine
-
- EGFR
-
- epidermal growth factor receptor
-
- ES
-
- Enrichment Score
-
- FBS
-
- fetal bovine serum
-
- FDR
-
- false discovery rate
-
- FITC
-
- fluorescein isothiocyanate
-
- FPKM
-
- fragments per kilobase of exon per million reads mapped
-
- GATK
-
- Genome Analysis Toolkit
-
- GEPIA
-
- Gene Expression Profiling Interactive Analysis
-
- GISTIC
-
- Genomic Identification of Significant Targets in Cancer
-
- GO
-
- Gene Ontology
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- H&E
-
- hematoxylin and eosin
-
- HCC
-
- hepatocellular carcinoma
-
- HRP
-
- Horseradish Peroxidase
-
- IgG
-
- immunoglobin G
-
- IGV
-
- Integrative Genomics Viewer
-
- IHC
-
- immunohistochemistry
-
- IP
-
- immunoprecipitation
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- lncRNA
-
- long non-coding RNA
-
- LR
-
- early apoptotic cells
-
- LUAD
-
- lung adenocarcinoma
-
- m5C
-
- 5-methylcytosine
-
- m6A
-
- N6-methyladenosine
-
- MB
-
- methylene blue
-
- MeRIP
-
- methylated RNA immunoprecipitation
-
- MeRIP-qPCR
-
- methylated RNA immunoprecipitation -qPCR
-
- MeRIP-seq
-
- methylated RNA immunoprecipitation sequencing
-
- MTT
-
- 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromid
-
- NC
-
- negative control.
-
- NJLCC
-
- Nanjing Lung Cancer Cohort
-
- NSCLC
-
- non-small-cell lung cancer
-
- OS
-
- overall survival
-
- PBS
-
- phosphate belanced solution
-
- PMSF
-
- phenylmethanesulfonyl fluoride
-
- PVDF
-
- polyvinylidene difluoride
-
- qRT-PCR
-
- quantitative real-time polymerase chain reaction
-
- RIP
-
- RNA immunoprecipitation
-
- RIPA
-
- Radio-Immunoprecipitation Assay
-
- RPMI
-
- Roswell Park Memorial Institute
-
- siRNA
-
- small interfering RNA
-
- SPSS
-
- Statistical Product and Service Solutions
-
- SSC
-
- Saline-Sodium Citrate
-
- STR
-
- short tandem repeat
-
- TCGA
-
- The Cancer Genome Atlas
-
- TPM
-
- transcripts per million
-
- UR
-
- late apoptotic cells
-
- UV
-
- ultraviolet
1 BACKGROUND
Non-small-cell lung cancer (NSCLC) is the primary type of lung cancer and is the leading cause of cancer mortality worldwide [1]. LUAD is the most prevalent subtype of NSCLC. Over the past years, its morbidity and mortality rate has increased worldwide because LUAD is mostly diagnosed at a late stage, limiting clinical treatment options [2]. Thus, LUAD diagnosis and treatment remain major challenges, which rely on a better understanding of the tumorigenesis mechanisms of LUAD [3].
Accumulating evidence has revealed that genetic, epigenetic and transcriptomic alterations could participate in the tumorigenesis of LUAD [4-6]. Strikingly, epigenetic regulation may lead to profound gene expression changes to facilitate LUAD formation and development [7]. Traditionally, epigenetic alterations included DNA methylation and histone modification [8]. Recent studies have shown that RNA modifications, as important epigenetic markers, are involved in regulating many biological processes, including tumorigenesis [9, 10]. For instance, METTL3, as an m6A methyltransferase, could promote the translation of epidermal growth factor receptor (EGFR) and the Hippo pathway effector TAZ in human cancer cells [11]. NSUN2, as an RNA methyltransferase, mediates the 5-methylcytosine (m5C) modification of H19 long non-coding RNA (lncRNA) to promote the occurrence and development of hepatocellular carcinoma (HCC) [12]. At the RNA level, an increasing number of chemical modifications have been discovered. Among them, as a posttranscriptional modification, m6A RNA methylation is the most prevalent chemical modification at the RNA epigenetic level [13-15], which regulates gene expression by influencing transcript stability, splicing, translation efficiency and cap-independent translation [16]. Generally, the m6A methyltransferase complex deposits m6A modifications on RNAs, forming a protein complex consisting of an RNA methyltransferase (METTL3, METTL14, WTAP, and KIAA1429 [also known as VIRMA], termed “writers”) and recognized by YTH m6A RNA binding proteins 1-3 (YTHDF1, YTHDF2 and YTHDF3, an important part of “readers”) [17, 18]. Recent evidence suggests that m6A modification is associated with cancer proliferation, invasion, and metastasis [11, 19, 20]. “Writers” (RNA methyltransferases), as a key member of m6A-related enzymes, are essential for m6A methylation. Several researchers have found that RNA methyltransferases have dysregulated expressions and play important roles in tumorigenesis [21]. For example, METTL3 is upregulated in human bladder cancer compared to normal tissues and promotes bladder cancer progression via the AFF4/NF-κB/MYC signaling network [22]. WTAP is highly expressed in HCC tissues and promotes liver cancer development via the HuR-ETS1-p21/p27 axis [23].
Although these studies indicate that RNA methyltransferases could play important roles in tumorigenesis, few have explored the reason for the activation of m6A methyltransferase in tumorigenesis, especially at the genome level, such as copy number variations (CNVs). In this study, we aimed to uncover the activated reason of RNA methyltransferase KIAA1429, and we further investigated its biological function and molecular regulatory mechanism in tumorigenesis of LUAD.
2 METHODS
2.1 The copy number variations of m6A methyltransferases
First, based on the whole-genome sequencing data of 55 LUAD patients in the Nanjing Lung Cancer Cohort (NJLCC) database (https://ega-archive.org/datasets/EGAD00001004071), the sample genome copy number fragment data were identified and obtained using the Genome Analysis Toolkit (GATK) 4 process. In addition, we also obtained the copy number data of LUAD samples (a total of 511 LUAD samples containing copy number variations information) from The Cancer Genome Atlas (TCGA) database by downloading from https://gdac.broadinstitute.org/. Combined with the human reference genome data, Genomic Identification of Significant Targets in Cancer (GISTIC) 2.0 software was used to identify gene copy number variations. GISTIC analysis was performed using the GISTIC 2.0 pipeline (GenePattern, https://genepattern.broadinstitute.org/). The corresponding GISTIC score = −2, ≤ −1, ≥ 1, and = 2 represented a gene undergoing homozygous deletion, copy-number loss, copy-number gain, and amplification, respectively [24]. Then, we calculated the copy amplification ratio for each m6A RNA methyltransferase. By combining transcriptome sequencing data obtained from the NJLCC and TCGA databases, we used Spearman rank-sum test to evaluate the association of expression level with the copy number of KIAA1429 in samples. The copy numbers of KIAA1429 in the cells were determined using the threshold cycle values from the respective standard curves.
2.2 Cell culture
The LUAD cell lines used in this study, including A549, SPCA1, NCI-H1299 and the normal human bronchial epithelial cell line HBE, were purchased from the Cell library of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). A549 cell line was cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Scientific HyClone, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA, USA). SPCA1, NCI-H1299 and HBE cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific Gibco, Carlsbad, CA, USA) supplemented with 10% FBS. All cell lines were identified by short tandem repeat (STR) analysis (Biowing, Shanghai, China). The cells were cultured in an incubator (Thermo Scientific, Carlsbad, CA, USA) at a temperature of 37°C and CO2 concentration of 5%.
2.3 Transfection
The nucleotide sequences of si-KIAA1429/si-YTHDF2/si-BTG2 are shown in Supplementary Table S1. The negative control small interfering RNA (siRNA) was obtained from Invitrogen (San Francisco, CA, USA). Transfections were performed using the Lipofectamine 2000 Kit (Invitrogen, San Francisco, CA, USA). Typically, the cells were evenly added to 6-well culture plates at a certain concentration and transfected with siRNA on the next day. Lentivirus vectors of KIAA1429 knockdown were obtained from GeneChem (Shanghai, China). LUAD cells were added to 6-well dishes and transfected with KIAA1429 knockdown lentivirus or scramble control. KIAA1429 and BTG2 complementary DNA (cDNA) were synthesized and cloned into the expression vector pcDNA3.1. Following the manufacturer's instructions, the plasmid was transfected into LUAD cells using the X-tremeGENE™ HP DNA Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA).
2.4 RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR) analyses
TRIzol reagent was used to lyse A549, SPCA1, NCI-H1299 and HBE cells and extract RNA. Then, the RNA was reversely transcribed into cDNA using a reverse transcription kit (Takara, Beijing, China). TB Green (Takara, Beijing, China) was used to perform qRT-PCR assays according to the manufacturer's instructions. The results were normalized with GAPDH. The primers used are shown in Supplementary Table S1. Each assay was replicated three times.
2.5 Cell proliferation assays
A cell viability assay kit was performed using the Cell Proliferation Reagent Kit I (3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, MTT) (Roche Applied Science, Indianapolis, IN, USA). The cells were seeded in 96-well plates at 2×103 cells/well. After transfection for 24 hours, cell viability was determined every 24 hours following the manufacturer's protocol. Each assay was replicated three times. A 5-ethynyl-2’-deoxyuridine (EdU) cell proliferation detection kit (RIBOBIO, Guangzhou, China) was used to detect the cell proliferation ability. The cells were cultured in 24-well plates at 5×103 cells/well. After 48 hours of transfection, 50 mmol/L EdU labeling medium was added and then incubated for 2 hours at 37°C and 5% CO2. Using 4% paraformaldehyde and 0.5% Triton X-100, the cells were stained with an anti-EdU working solution, and the cell nuclei were labeled with 4’,6-diamidino-2-phenylindole (DAPI) (RIBOBIO, Guangzhou, China). The percentage of EdU-positive cells was analyzed using fluorescence microscopy (Carl Zeiss, Jena, Germany). Each assay was replicated three times.
2.6 Colony formation assays
In the colony formation experiment, 1×103 cells/well were added into a 6-well dish and cultured in the medium for approximately two weeks. The cells were first fixed with methanol (Sigma Aldrich, St. Louis, MO, USA) and then stained with 0.1% crystal violet (Beyotime Biotechnology, Shanghai, China) for 15 minutes. The colony-forming ability of the clones was reflected by comparing the number of colonies in each group. Each assay was replicated three times.
2.7 Wound healing assays
The cells were seeded in 6-well dishes with a culture medium overnight. Before transfection, a 10-μL sterile plastic tip was used to create a wound line across the surface of each dish. Then, the suspension cells were removed with phosphate balanced solution (PBS) (Thermo Fisher Scientific Gibco, Carlsbad, CA, USA), and a new culture medium was added in a humidified 5% CO2 incubator at 37°C. Images were taken with a microscope (Olympus, Tokyo, Japan) at 24 hours. Each assay was replicated three times.
2.8 Cell migration assays
The transfected cells in serum-free medium and the medium with 10% FBS were placed in the upper chamber (Corning, NewYork, NY, USA) and the lower chamber, respectively. After 24 hours of incubation, the cells that passed through the membrane were fixed and stained with methanol and 0.1% crystal violet and then photographed and counted using an inverted microscope (Olympus, Tokyo, Japan). The experiment included three replicates.
2.9 Flow cytometric analysis
After two days of transfection, the medium supernatant was first collected into a flow tube (BD Bioscience, East Rutherford, NJ, USA), and the cells were collected by trypsinization (Thermo Fisher Scientific Gibco, Carlsbad, CA, USA). Double staining with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide was performed using the FITC Annexin V Apoptosis Detection Kit (BD Bioscience, East Rutherford, NJ, USA) based on the manufacturer's instructions. The cells were analyzed with a flow cytometer (FACScan, BD Bioscience, East Rutherford, NJ, USA) equipped with Cell Quest software (BD Bioscience, East Rutherford, NJ, USA). The cells were divided into viable cells, dead cells, early apoptotic cells, and late apoptotic cells. The relative proportion of early apoptotic cells and late apoptosis cells were recorded and compared with the control group. Each assay was replicated three times.
2.10 Western blot assay and antibodies
The treated cells were lysed using Radio-Immunoprecipitation Assay (RIPA) buffer (Beyotime Biotechnology, Shanghai, China) supplemented with a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA, dilution ratio, 1:100) and phenylmethanesulfonyl fluoride (PMSF) (Roche Applied Science, Indianapolis, IN, USA, dilution ratio, 1:1000). A 10% precast gel (SurePAGE™, Nanjing, China, 10 wells) was used to load the lysed protein. The protein was then transferred to polyvinylidene difluoride (PVDF) membranes (Sigma Aldrich, St. Louis, MO, USA), blocked with bovine serum albumin (BSA) (Thermo Scientific, Carlsbad, CA, USA), and incubated overnight with primary antibody. The membrane was washed with wash buffer and probed with a secondary antibody (Beyotime Biotechnology, Shanghai, China). Exposure records and quantification were carried out by a developing instrument (Bio-Rad, Universal Hood II, CA, USA). GAPDH antibody was purchased from Cell Signaling Technology (Danvers, MA, USA) (dilution ratio, 1:1000, 5174S) and used as a control. Anti-KIAA1429 (dilution ratio, 1:1000, ab271136) and anti-BTG2 (dilution ratio, 1:200, ab197362) were purchased from Abcam (Cambridge, UK). Each assay was replicated three times.
2.11 Transcriptome sequencing
Total RNA was isolated from KIAA1429 knockdown and control A549 cells. The concentration of each sample was measured using a NanoDrop 2000 (Thermo Scientific, Carlsbad, CA, USA). The quality was evaluated by an Agilent 2200 (Agilent, Palo Alto, CA, USA). The sequencing library of each RNA sample was prepared using the Ion Proton Total RNA-Seq Kit version 2 based on a protocol provided by the manufacturer. The corresponding data are listed in Supplementary Table S2. Each assay was replicated three times.
2.12 Gene ontology (GO) analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Version 6.8 Beta) was used to perform the enrichment analysis, an online tool for GO enrichment analysis (https://david.ncifcrf.gov/) [25]. The GO biological process terms were selected from the top significantly enriched clusters. Bonferroni-corrected P-values were identified as significant when below 0.05.
2.13 Gene set enrichment analysis (GSEA)
To determine the differentially expressed gene (DEG) response to KIAA1429 knockdown, we used GSEA in the GSEA tool to list particular pathways described in previously known gene sets. A normalized enrichment score was calculated to compare enrichment analysis results across gene sets [26, 27].
2.14 RNA m6A dot blot assays
Total RNA was first loaded onto a nitrocellulose membrane (Amersham, GE Healthcare, USA) installed in a BioDot apparatus (Bio-Rad, Hercules, CA, USA) with ice-cold 20×Saline-Sodium Citrate (SSC) buffer (Sigma Aldrich, St. Louis, MO, USA). Then, the membrane was cross-linked using ultraviolet (UV), blocked, incubated with m6A antibody (202003, 1:2000, Synaptic Systems, Goettingen, SN, Germany) overnight at 4°C and subsequently incubated with Horseradish Peroxidase (HRP)-conjugated goat anti-mouse immunoglobin G (IgG) (Beyotime Biotechnology, Shanghai, China) for 1 hour. Finally, the membrane was visualized by an imaging system (Bio-Rad, Universal Hood II, CA, USA). The membrane was stained with 0.02% methylene blue (MB) (Beyotime Biotechnology, Shanghai, China) in 0.3 mol/L sodium acetate (pH 5.2) (Sigma Aldrich, St. Louis, MO, USA) to ensure the consistency of the baseline between the different groups. Experiments were repeated three times.
2.15 Methylated RNA immunoprecipitation sequencing (MeRIP-seq)
Total RNA was isolated as before, and purified RNA fragments were isolated using the RiboMinus™ Eukaryote Kit version 2 (Invitrogen, Waltham, MA, USA). The fragmented RNA was sorted into two portions. First, immunomagnetic beads with premixed m6A antibody were added to enrich the mRNA fragments containing m6A methylation. The other was used as a control to construct a conventional transcriptome sequencing library. Library preparation and high-throughput sequencing were performed by the Illumina Nova seq 6000 platform of LC-BIO Biotech (Hangzhou, China). Then, the analysis results of enrichment of m6A peaks and location results were converted into bw files and imported into the Integrative Genomics Viewer (IGV) software (Los Angeles, CA, USA) for visual display. According to the position of RNA modification sites of m6A and total reads number in the bw file, the number of reads obtained from the immunoprecipitation (IP) library and Input library was used to quantify the degree of m6A modification. This data was reflected in the coordinate interval of each modification to form a visual representation in IGV software. Each assay was replicated three times.
2.16 Methylated RNA immunoprecipitation (MeRIP) assay and qRT-PCR
Total RNA was isolated from stable KIAA1429 knockdown A549 cells and controls. Chemically fragmented RNA (∼100 nt) was incubated with an m6A antibody for immunoprecipitation according to the standard protocol of the Magna MeRIP™ m6A Kit (Merck Millipore, Billerica, MA, USA). Briefly, total RNA was preheated at 94°C for 10 minutes, Ethylene Diamine Tetraacetic Acid (EDTA) (Sigma Aldrich, St. Louis, MO, USA) was immediately added, and 3mol/L sodium acetate (Sigma Aldrich, St. Louis, MO, USA), glycogen (Sigma Aldrich, St. Louis, MO, USA) and 100% ethanol (Sigma Aldrich, St. Louis, MO, USA) were added, and the samples were incubated at −80°C overnight. The next day, magnetic beads were prepared, and the MeRIP reaction mixture was prepared and incubated with m6A antibody at 4°C for 2 hours. The last m6A RNA was eluted with 10 mg of m6A 5’-monophosphate sodium salt (Merck Millipore, Billerica, MA, USA) at 4°C for 1 hour. Enrichment of m6A was analyzed using qRT-PCR. Experiments were repeated three times.
2.17 Tumor formation assay in mice model
Five-week-old male athymic BALB/c mice were purchased from GemPharmatech (Nanjing, China) and maintained under specific pathogen-free conditions. For the in vivo cell proliferation assay, A549 and SPCA1 cells were stably transfected with sh-Ctrl and sh-KIAA1429 using lentivirus (GeneChem, Shanghai, China). The cells were subcutaneously injected into either side of the posterior flanks of the mouse. The tumor volume was measured every few days (length×width2×0.5). At the end of the experiment, the mice were euthanized, and the tumors were removed, weighed, and then fixed for hematoxylin and eosin (H&E), BTG2 and Ki-67 immunostaining analysis. To observe tumor metastasis in the lungs, treated SPCA1 cells were injected into the mice's tail vein. After 60 days, the mice were euthanized, and the lungs were removed. Images of the lungs were taken for recording and used for H&E immunostaining analysis, and the nodules on the lung were counted. All animal care procedures and experiments were conducted following animal welfare guidelines and based on the “3Rs” principle (reduce, replace, refine). This study was conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Our protocol was approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University.
2.18 RNA stability assays
After siRNA and plasmid transfection of A549 and SPCA1 cells for 24 hours, the cells were treated with 10 μg/mL actinomycin D (Sigma Aldrich, St. Louis, MO, USA), then collected at 0, 2, and 4 hours after treatment. Total RNA was extracted and detected by qRT-PCR. Each assay was replicated three times.
2.19 RNA immunoprecipitation (RIP) assays
The RIP experiment was performed using the Magna RIP™ RNA binding protein immunoprecipitation kit (CAT.17-701, Millipore, Billerica, MA, USA), and all operations were performed following the manufacturer's instructions. The antibody against YTHDF2 for the RIP assay was purchased from Proteintech (24744-1-AP, Rosemont, PA, USA). Experiments were repeated three times.
2.20 Statistical analysis
All statistical analyses were performed using the Statistical Product and Service Solutions (SPSS) 22.0 software (SPSS Inc., Chicago, IL, USA). The statistical significance of differences between experimental groups and controls was assessed by the one-sample t-test, and two-tailed unpaired or paired t-test. One-way Analysis of Variance (ANOVA) followed by post hoc Tukey's test was used to compare multiple groups. Spearman's rank correlation coefficient was used to measure the correlation between two continuous variables. The overall survival (OS), the time from diagnosis to last follow-up or last date known to be still alive, was calculated using the Kaplan-Meier method and log-rank test for comparison. Two-sided P-values of 0.05 were selected for statistical significance.
3 RESULTS
3.1 Copy number amplification could drive higher expression of KIAA1429 in LUAD
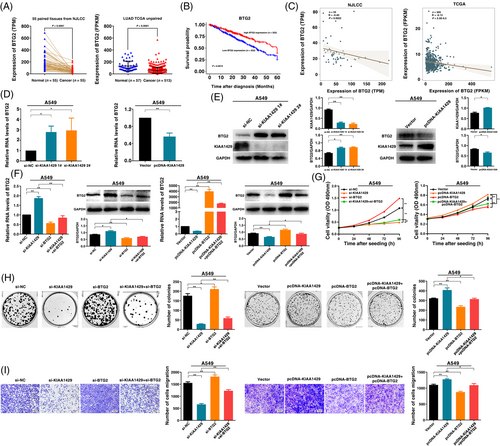
We analyzed the copy number amplification for m6A methyltransferases (METTL3, METTL14, WTAP, and KIAA1429) using whole genome sequencing and transcriptome sequencing of LUAD in the NJLCC (n = 55, these cases of the sample having both whole genome sequencing and transcriptome sequencing data) and TCGA database. We observed that KIAA1429 exhibited the highest copy amplification rate in LUAD samples (7.3% in the NJLCC database, 3.9% in the TCGA database) among the four main m6A methyltransferase genes (METTL3, METTL14, WTAP, and KIAA1429) (Figure 1A). Although METTL3 was also amplified in LUAD, the amplification rate was lower than KIAA1429. Many studies have examined the function of METTL3 in tumorigenesis, including lung cancer [11, 28]. Therefore, we focused on KIAA1429. Additionally, the expression of KIAA1429 was significantly correlated with the copy number level in both the NJLCC LUAD database and TCGA LUAD database (Figure 1B). We also examined the copy number amplification of KIAA1429 in A549, SPCA1, NCI-H1299 and HBE cell lines and found higher copy number of KIAA1429 in these three LUAD cell lines than in HBE (a normal human bronchial epithelial cell line) (Supplementary Figure S1A). Moreover, qRT-PCR analysis showed that the KIAA1429 expression was higher in diverse human LUAD cell lines than in normal human bronchial epithelial cell line (HBE) (Supplementary Figure S1B).
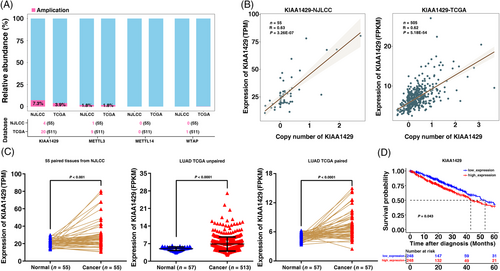
Next, we investigated KIAA1429 expression in 55 LUAD tumor tissues compared with their matched adjacent normal tissues in the NJLCC database. As shown in Figure 1C, the KIAA1429 expression level in cancer tissues was significantly higher than that in the corresponding normal tissues. We further validated this result in the TCGA data of LUAD and revealed that KIAA1429 was also overexpressed in unpaired and paired LUAD tissues (Figure 1C). We also found that high expression of KIAA1429 indicated a poor prognosis in the TCGA dataset (Figure 1D). However, low METTL3 expression revealed a significantly worse prognosis in LUAD patients (Supplementary Figure S1C). Other methyltransferases, including METTL14 and WTAP, were not statistically significant prognostic factors. The above results indicated that KIAA1429 expression was upregulated in tumor tissues compared with adjacent normal tissues and that high KIAA1429 expression was associated with poor prognosis.
3.2 KIAA1429 regulates LUAD cell proliferation and migration in vitro
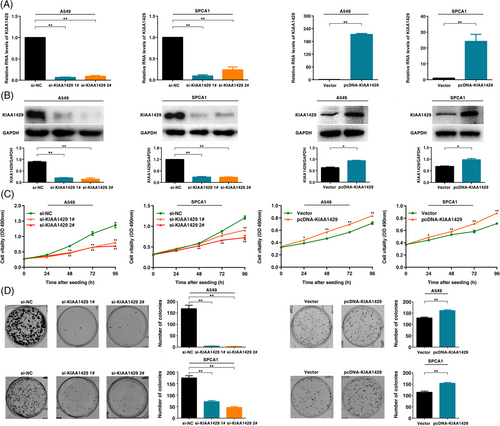
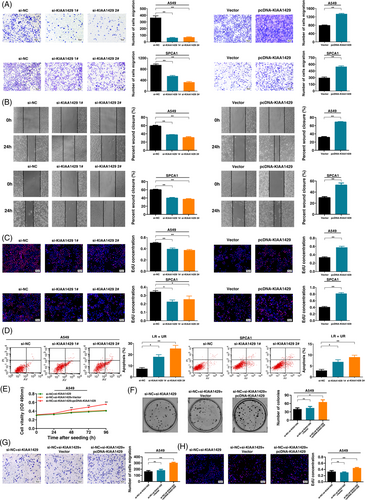
To explore the biological role of KIAA1429 in LUAD, we used LUAD cell lines (A549, SPCA1 and NCI-H1299). To confirm the function of KIAA1429, we used two independent siRNA-mediated knockdowns and plasmid-mediated overexpression. The mRNA and protein levels confirmed that the expression of KIAA1429 was successfully regulated (Figure 2A, B and Supplementary Figure S2A, B). MTT analysis showed that downregulation of KIAA1429 expression could significantly inhibit cell viability. In contrast, overexpression of KIAA1429 promoted cell proliferation (Figure 2C and Supplementary Figure S2C). Next, colony formation assays showed that the clone formation ability after knocking down KIAA1429 was significantly lower than that of the control group. Furthermore, overexpression of KIAA1429 increased the number of clones (Figure 2D and Supplementary Figure S2D). Transwell assays suggested that cell migration was inhibited after silencing KIAA1429. Overexpressed KIAA1429 promoted cell migration (Figure 3A and Supplementary Figure S2E). Similarly, compared to control cells, the migratory activity in wound-healing assays was reduced by knockdown of KIAA1429 expression in A549 and SPCA1 cells. Overexpression of KIAA1429 increased migratory activity (Figure 3B). Moreover, the results of the EdU assays demonstrated that KIAA1429 had a significant impact on LUAD cell proliferation (Figure 3C and Supplementary Figure S2F). Flow cytometric analysis found that knockdown of KIAA1429 significantly induced A549, SPCA1 and NCI-H1299 cell apoptosis (Figure 3D and Supplementary Figure S2G). Furthermore, we found that overexpression of KIAA1429 could partly reverse si-KIAA1429-mediated growth and migration suppression (Figure 3E-H). Collectively, the above results indicated that KIAA1429 played an oncogenic role in the tumorigenesis of LUAD.
3.3 KIAA1429 affected the overall m6A modification level and regulated the mRNA m6A levels of BTG2 in LUAD
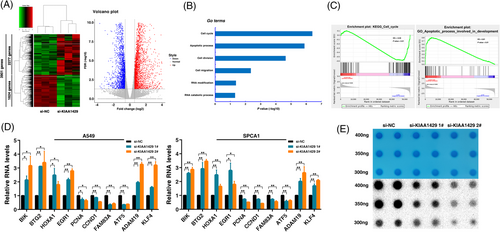
To delineate the functional implications of KIAA1429 and identify its downstream targets in LUAD, we performed RNA transcriptome sequencing to investigate the expression changes in KIAA1429 knockdown cells. In total, we identified 3901 differentially expressed genes (fold change ≥ 1.5), of which 2277 were upregulated and 1624 showed decreased expression in KIAA1429 knockdown A549 cells (Figure 4A and Supplementary Table S2). GO assay was used to explore the underlying pathways mediated by KIAA1429. The results revealed that differentially expressed genes were enriched in gene sets involved in the cell cycle, apoptotic process, cell migration, RNA modification and catabolic process (Figure 4B). Moreover, GSEA revealed that the gene sets were significantly related to the cell cycle and apoptotic processes involved in development (Figure 4C). qRT-PCR assays were performed to verify the changes in several differentially expressed mRNAs involved in growth, apoptosis, oncogenesis pathways and m6A pathway enrichment genes based on the results of RNA sequencing. As shown in Figure 4D, after KIAA1429 knockdown, BIK, BTG2, HOXA1, EGR1, ADAM19 and KLF4 showed increased expression, while the expression of PCNA, CCND1, FAM83A, and ATF5 was downregulated in both A549 and SPCA1 cell lines. Importantly, the m6A dot blot assay showed that the overall m6A level decreased after suppressing KIAA1429 expression, indicating that KIAA1429 could affect the overall m6A modification level in A549 cells working as an RNA methyltransferase (Figure 4E). Collectively, our data demonstrated that KIAA1429 might affect downstream target gene expression by regulating m6A modification.
Among the potential targets of KIAA1429, special attention was given to the BTG2 gene because it was significantly upregulated after the knockdown of KIAA1429. In addition, previous studies have found that as a tumor suppressor, BTG2 is involved in the tumorigenesis of multiple tumors [29-31]. In addition, the promoter DNA methylation level of BTG2 could be a stable and reliable biomarker for early-stage lung cancer [32]. Our results indicated that the expression of BTG2 in LUAD tissues was significantly lower than that of adjacent tissues in the NJLCC database and TCGA database (Figure 5A). We also found that low BTG2 expression was associated with poor prognosis in the TCGA dataset (Figure 5B). Further research suggested a negative correlation between KIAA1429 and BTG2 in the NJLCC and TCGA databases (Figure 5C). We found that knockdown of KIAA1429 indeed upregulated BTG2 expression, while overexpression of KIAA1429 inhibited BTG2 expression at the RNA and protein levels (Figure 5D, E and Supplementary Figure S3A, B). We found that overexpression of KIAA1429 could partly reverse si-KIAA1429-mediated BTG2 expression induction (Supplementary Figure S3C). Further results indicated that the knockdown of BTG2 could reverse KIAA1429 knockdown-mediated BTG2 induction at both the mRNA and protein levels. In contrast, overexpressed BTG2 reversed KIAA1429 overexpression-mediated BTG2 inhibition (Figure 5F, Supplementary Figure S3D-F). Functionally, knockdown of BTG2 promotes cell proliferation and migration, whereas overexpression of BTG2 inhibits cell proliferation and migration. In addition, the knockdown of BTG2 reversed KIAA1429 knockdown-mediated growth and migration inhibition. Overexpressed BTG2 partly reversed KIAA1429 overexpression-mediated growth and migration promotion (Figures 5G-I and Supplementary Figure S3G-I). In conclusion, KIAA1429 could regulate the expression of BTG2 to promote tumorigenesis of LUAD.
3.4 KIAA1429 mediates the expression of BTG2 in an m6A-YTHDF2-dependent manner
Combining the above results, we speculated that KIAA1429 might suppress BTG2 through m6A modification, thus promoting tumorigenesis in LUAD. To further verify the mechanism between KIAA1429 and BTG2, we performed MeRIP-seq assays and found a consistently decreased m6A level in the 3’untranslated region (3’UTR) of BTG2 after KIAA1429 knockdown (Figure 6A). Then, a methylated RNA immunoprecipitation-qPCR (MeRIP-qPCR) assay also verified the reduction in the m6A modification level of BTG2 after KIAA1429 knockdown (Figure 6B). Thus, we confirmed that KIAA1429 regulates the expression of BTG2 in an m6A modification-dependent manner.
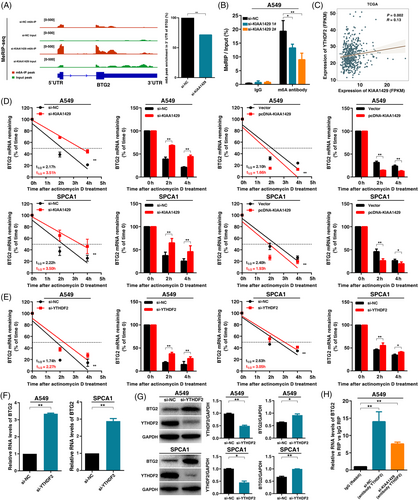
Next, we investigated how m6A affected the expression of BTG2. Previous researchers have found that m6A “reader” proteins, including YTHDF1 and YTHDF2, selectively recognize m6A modifications and mediate m6A-containing mRNA degradation, especially YTHDF2, which could lead to mRNA degradation [33]. These results led us to hypothesize that m6A-containing BTG2 mRNA might be recognized and bound by YTHDF2. To test this hypothesis, our analysis found a positive expression correlation between KIAA1429 and YTHDF2 (Figure 6C). To confirm the function of YTHDF2 in LUAD cells, we used siRNA-mediated knockdown of the expression of YTHDF2. MTT analysis showed that YTHDF2 knockdown inhibited A549 cell viability (Supplementary Figure S4A). EdU assays demonstrated that YTHDF2 knockdown inhibited the cell proliferative capacity of A549 (Supplementary Figure S4B). Colony formation assays showed that YTHDF2 knockdown inhibited A549 cell clone formation ability (Supplementary Figure S4C). Next, Transwell assays showed that cell migration was significantly lower after silencing YTHDF2 (Supplementary Figure S4D). To further verify the influence of KIAA1429 and YTHDF2 on the RNA stability of BTG2, we treated A549 and SPCA1 cells with the transcription inhibitor actinomycin D after knockdown or overexpression of KIAA1429. Indeed, we found that the knockdown of KIAA1429 increased the half-life of BTG2 transcripts. In contrast, overexpression of KIAA1429 promoted mRNA degradation of BTG2 (Figure 6D). The half-life of BTG2 transcripts was significantly increased after silencing YTHDF2 in both A549 and SPCA1 cells (Figure 6E). These results indicated that the m6A modification of BTG2 mRNA could be recognized and bound by YTHDF2. qPCR and Western assays showed that BTG2 mRNA and protein levels were significantly increased after the knockdown of YTHDF2 expression (Figure 6F and G). Moreover, the RIP assay showed that the YTHDF2 protein could directly bind to BTG2 mRNA. The binding abundance of YTHDF2 on BTG2 mRNA decreased significantly after silencing KIAA1429 (Figure 6H). Taken together, these results showed that KIAA1429 could regulate BTG2 in an m6A-YTHDF2-dependent manner in LUAD tumorigenesis.
3.5 KIAA1429 regulates LUAD cell proliferation and migration in vivo
To further determine whether KIAA1429 affects the proliferation of LUAD cells in vivo, A549 and SPCA1 cell lines stably transfected with sh-KIAA1429 were inoculated into mice using lentivirus. A few days after injection, compared with the control group, the volume of tumors in the sh-KIAA1429 group was significantly reduced (Figure 7A). Moreover, at the end of the experiment, the average tumor weight of the sh-KIAA1429 group was significantly lower than that of the control group (Figure 7A). The positive rate of Ki-67 staining for tumors formed by stable sh-KIAA1429-transfected A549 and SPCA1 cells was significantly lower than that of the control group (Figure 7A). The positive rate of BTG2 staining was higher in KIAA1429-depleted cells than in the control group (Figure 7A). These findings indicated that knockdown of KIAA1429 inhibited tumor growth in vivo.
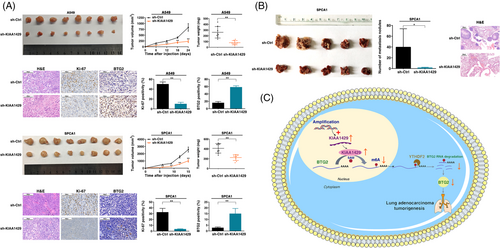
To verify the effect of KIAA1429 on cell transfer in vivo, sh-KIAA1429 and a control vector were stably transfected into SPCA1 cells, and the transfected cells were injected into the mice through the tail vein. After 60 days, the metastatic nodules on the lung surface were counted. Inhibition of KIAA1429 expression reduced the number of metastatic nodules compared with the control group (Figure 7B). The nodules were confirmed by inspecting the entire lungs, slicing the lung, and staining with H&E (Figure 7B). Therefore, the in vivo results supplemented the results of the functional in vitro studies of KIAA1429. In summary, these results suggest that gene amplification-driven KIAA1429 contributes to tumorigenesis of LUAD by regulating BTG2 in an m6A-YTHDF2-dependent manner (Figure 7C).
4 Discussion
This study found that KIAA1429 had high copy number amplification in LUAD based on analyses from the NJLCC and TCGA datasets and was associated with poor prognosis in the TCGA dataset. In vitro and in vivo experiments showed that KIAA1429 regulated the cell proliferation and migration, and affected the global m6A modification level of LUAD cells. Moreover, we confirmed that KIAA1429 could affect the expression of BTG2 by regulating the stability of BTG2 mRNA in an m6A-YTHDF2 dependent way, thus promoting the tumorigenesis of LUAD.
Prior reports showed that RNA methylation has the same characteristics as epigenetic DNA and histone modifications. Among these modifications, m6A is the most prevalent internal modification in polyadenylated mRNAs [34]. As a “writer” of m6A, RNA methyltransferase is an important factor in aberrant m6A modification. Previous studies noted that RNA methyltransferase was abnormally expressed in various malignant tumors [28, 35]. However, the upstream mechanism of dysregulation has rarely been reported. Our study systematically analyzed the copy number changes of m6A “writers” (KIAA1429, METTL3, METTL14 and WTAP) in LUAD. We found that KIAA1429 and METTL3 had copy amplification in LUAD samples. Although METTL3 was amplified in LUAD, the amplification rate of KIAA1429 was higher than that of METTL3. Many studies have focused on the function of METTL3 in tumorigenesis [18, 28, 35]. As an m6A “writer”, KIAA1429 recruits the catalytic core components METTL3/METTL14/WTAP to guide m6A modification [17, 36]. KIAA1429 mediates preferential mRNA methylation in the 3’UTR and near the stop codon [37]. Previous studies have shown that KIAA1429 could play an essential role in tumorigenesis, including lung cancer [38-40]. For instance, KIAA1429 promotes the migration and invasion of HCC by inhibiting ID2 via upregulating m6A modification of ID2 mRNA [38]. KIAA1429 regulates the expression of MUC3A by modifying m6A, thus regulating the proliferation and migration of LUAD cells [39]. KIAA1429 can accelerate the gefitinib resistance of NSCLC by targeting HOXA1 3’UTR to enhance its mRNA stability [40]. However, the reason for the activation of KIAA1429 remains unclear. Our study first discovered that the activation of KIAA1429 was partly due to copy number amplification in LUAD. These results indicated that genome copy number amplification might be involved in abnormal RNA methylation modification in tumorigenesis. We also found that KIAA1429 was significantly highly expressed in LUAD cells and tissues and that high expression of KIAA1429 was associated with poor prognosis of LUAD patients, which was consistent with the results of previous studies, indicating that the expression level of KIAA1429 could serve as a prognostic marker for LUAD patients [41-43]. We functionally demonstrated the essential role of KIAA1429 in promoting LUAD growth and metastasis via in vitro and in vivo assessments. Further study showed that KIAA1429 transcriptionally regulated a large set of genes linked to cell proliferation and cell migration in LUAD cells, such as BIK, BTG2, HOXA1, EGR1, PCNA, CCND1, FAM83A and ATF5, and are thus involved in the development of LUAD.
We also demonstrated that KIAA1429 acted as an m6A “writer” to affect the global m6A level in LUAD cells. Then, we conducted RNA-seq in A549 cell lines and found that BTG2 was a target gene of KIAA1429. BTG2 is a member of the B cell translocation gene (BTG) family and can regulate cell cycle progression, apoptosis, and differentiation [31]. Several reports have shown that BTG2 could restrain the cell growth and migration of multiple cancers, including lung cancer [44-46]. Moreover, BTG2 is one of the p53 target genes involved in the DNA damage repair process in tumor cells [47, 48]. In addition, previous studies have shown that BTG2 was downregulated in many tumor types, including lung cancer, mainly because it could be modified by DNA methylation in the promoter [48, 49]. However, we found that RNA methylation modification (m6A) could also be an epigenetic marker that inhibits the expression of BTG2 in LUAD. Here, we identified that the tumor suppressor BTG2 was significantly downregulated in LUAD tissues and demonstrated a negative correlation with KIAA1429. We also found BTG2 to be a direct downstream target of methyltransferase KIAA1429-mediated m6A modification. Taken together, we discovered that the global genes were regulated by KIAA1429, suggesting that KIAA1429 may have a carcinogenic effect on the progression of LUAD. Our results suggested that higher expression of KIAA1429 promotes LUAD tumorigenesis by regulating the expression of critical tumor suppressor genes at the RNA epigenetic level, which added a new layer of epigenetic alteration that contributed to the development of LUAD.
The influences of m6A modification on mRNA are mediated by specific m6A binding proteins, known as m6A “readers” [9]. The YTH domain family protein YTHDF1-3 was identified as an m6A-binding protein in eukaryotic mammals [33, 50]. YTHDF2 recognizes and destabilizes m6A-containing mRNA, and YTHDF1 and YTHDF3 modulate mRNA translation and promote protein synthesis [51]. Because the YTH domain family could play regulatory roles in various biological processes, we hypothesize that m6A-binding proteins might play important roles in the m6A modification mediated by KIAA1429. In our study, we found that the m6A “writer” KIAA1429 and the m6A “reader” YTHDF2 could both modulate BTG2 expression by regulating its mRNA stability. Therefore, we provided evidence showing that KIAA1429 could mediate the expression of BTG2 in an m6A-YTHDF2-dependent manner, thereby promoting LUAD tumorigenesis.
5 CONCLUSIONS
In summary, we revealed that copy number amplification could drive higher expression of RNA methyltransferase KIAA1429 in LUAD, and KIAA1429 could promote the proliferation and metastasis of LUAD cells in vitro and in vivo. As an RNA methyltransferase gene, KIAA1429 functions in LUAD by regulating the mRNA of tumor suppressor gene BTG2 in an m6A-YTHDF2-dependent manner, affecting its mRNA stability, thereby promoting LUAD tumorigenesis. Overall, our study suggests that KIAA1429 played an important role in the tumorigenesis of LUAD and could be regarded as a potential and promising therapeutic target for LUAD.
DECLARATIONS
ACKNOWLEDGMENTS
This work was supported by the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (81521004), the National Natural Science Foundation of China (81922061, 82172992, 81903391, 81702266), the Research Unit of Prospective Cohort of Cardiovascular Diseases and Cancers, Chinese Academy of Medical Sciences (2019RU038), Natural Science Foundation of Jiangsu Province (BK20211253, BK20190148), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_0195).
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
CZ, QS and XZ: contributed to designing and organizing the experiments and writing the manuscript. NQ, ZNP, MZ, CW, NL and JCD: contributed to carrying out the data analysis, data analysis. YYG and CWY: contributed to the execution and supervision of experimental operations. GFJ, HXM, ZBH: contributed to design of the work. EBZ, FWT and HBS: contributed to conceiving the ideas, supervising the study, and writing the manuscript. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the Human Research Ethics Committee of Nanjing Medical University (2017-030), and all of the participants signed an informed consent form. The study protocol was approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University (IACUC-1812039).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.




