Matrisomal genes in squamous cell carcinoma of head and neck influence tumor cell motility and response to cetuximab treatment
Abbreviations
-
- AUC
-
- area under the curve
-
- CAF
-
- cancer-associated fibroblast
-
- CTX
-
- cetuximab
-
- DFI
-
- disease-free interval
-
- DSS
-
- disease-specific survival
-
- ECM
-
- extracellular matrix
-
- EGFR
-
- epidermal growth factor receptor
-
- HNSCC
-
- head and neck squamous cell carcinoma
-
- HPV
-
- (human papillomavirus)
-
- HR
-
- hazard ratio
-
- IC50
-
- half-maximal inhibitory concentration
-
- MMP
-
- matrix metalloproteinase
-
- mRNA
-
- messenger ribonucleic acid
-
- MTT
-
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
-
- PCR
-
- polymerase chain reaction
-
- PD-1
-
- programmed cell death protein 1
-
- R/M HNSCC
-
- recurrent/metastatic head and neck squamous cell carcinoma
-
- ROC
-
- receiver operating characteristics
-
- siRNA
-
- small interfering ribonucleic acid
-
- TCGA-HNSC
-
- the cancer genome atlas-head and neck squamous cell carcinoma
-
- TMI
-
- tumor matrisome index
Dear Editor,
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer globally [1, 2]. Despite the improvement in treatment modalities, up to 50% of HNSCC patients still develop recurrent/metastatic (R/M) disease [3], and platinum-based chemotherapy with cetuximab and/or pembrolizumab has become the standard of care [4]. However, R/M HNSCC is challenging to treat, the prognosis is poor, and there is an unmet need for new therapeutic targets.
The extracellular matrix (ECM) has emerged as a favorable target for cancer therapy [5]. The ECM proteins are dysregulated and derived from cancer cells to promote tumor growth and expansion [6]. We previously reported a 29-gene tumor matrisome index (TMI) that impacts prognosis and predicts the clinical outcomes of 11 cancer types [7, 8]. The 29 genes comprised of collagens, glycoproteins, proteoglycans, matrix metalloproteinases (MMP), secreted factors, and ECM-affiliated proteins. This TMI has not been explored in HNSCC, and how individual components contribute to tumor development is not well-understood in HNSCC. In this work, we examined the prognostic application of TMI in HNSCC and the influence of matrisome components MMP1 (interstitial collagenase) and MMP12 (macrophage elastase) on the metastatic potential of HNSCC lines and the response to platinum and cetuximab therapy. The experimental details are provided in the Supplementary Materials and Methods, including the characteristics of patient-derived HNSCC cell lines (Supplementary Table S1) and primer sequences used for expression quantification in quantitative real-time-polymerase chain reaction (qRT-PCR) experiments (Supplementary Table S2).
Using the expression array data for a cohort of cases with squamous cell carcinoma of the tongue [9], we found that tumor tissues had a higher TMI than normal tissues (Figure 1A), consistent with prior reported data in other cancers [7]. Using the receiver operating characteristics (ROC) curve, TMI could distinguish cancers from normal tissues with an area under the curve (AUC) of 0.970 (Figure 1B), demonstrating promising diagnostic utility. Distinct TMI values were observed across different subpopulations of HNSCC, including HNSCC1 (a mixture of locally advanced and earlier stage), HNSCC2 (metastatic), and HNSCC3 (locally advanced) (Figure 1C). Importantly, the TMI stratified tongue carcinoma patients into low- and high-risk groups based on the risk of relapse (Figure 1D), potentially allowing closer follow-up of patients at high risk. The performance of TMI across diagnosis and stage was further validated in The Cancer Genome Atlas-Head and Neck Squamous Cell Carcinoma (TCGA-HNSC) dataset corroborating the significance of TMI in patient stratification (Supplementary Figure S1A-D).
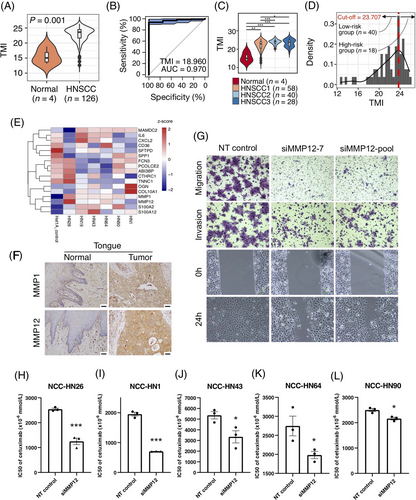
Effect of matrisomal genes on prognosis, tumor metastasis and drug response in HNSCC. (A-D) Diagnostic performance of TMI and patient stratification using patient-derived tongue carcinoma data. P-values were assessed by the Mann-Whitney-Wilcoxon test and the number of samples (n) are stated. (A) TMI of normal and tumor tissues. (B) The area under the ROC curve (AUC) of the TMI. The AUC value and the optimal cut-off are stated. (C) TMI of normal and subpopulations of tumor tissues. HNSCC1 (a mixture of locally advanced and earlier stage), HNSCC2 (metastatic), HNSCC3 (locally advanced). (D) Distribution of TMI and patient stratification. The optimal cut-off of TMI is stated. (E) Heatmap representing mRNA expression of TMI-comprising genes in six patient-derived HNSCC lines and a normal squamous epithelial line Het1A. The mRNA expression was normalized to beta-actin and the log2 fold change was calculated relative to Het1A. MMP1 and MMP12 were robustly upregulated across all HNSCC lines. (F) Immunohistochemistry staining of MMP1 and MMP12 in paired tongue tumor and normal tongue tissue. MMP1 and MMP12 were overexpressed in the tongue tumor. Scale bar represents 50μm. (G) siRNA against MMP12 significantly reduced the migration, invasion and wound healing ability relative to control in NCC-HN26 cells. (H-L) IC50 values in response to cetuximab. MMP12 knockdown improved the sensitivity of HNSCC cells to cetuximab. Error bars represent standard error mean (SEM). The asterisks represent statistical significance (***: P ≤ 0.001, **: P ≤ 0.01, *: P ≤ 0.05, ns: P > 0.05).
Abbreviations: HNSCC, head and neck squamous cell carcinoma; TMI, tumor matrisome index; ROC, receiver operating characteristic; AUC, area under the curve; mRNA, messenger ribonucleic acid; siRNA, small interfering ribonucleic acid; IC50, half-maximal inhibitory concentration; SEM, standard error mean
The TMI maintained an excellent diagnostic performance, irrespective of transcriptomics profiling platforms. Five machine learning models were trained using the microarray-derived expression profile of TMI-comprising genes from the tongue carcinoma dataset and tested on the RNA-seq-derived TCGA-HNSC dataset (Supplementary Figure S1E-G, Supplementary Table S3). The logistic regression model achieved the highest AUC of 0.965 in differentiating HNSC from normal tissues, while the remaining models had AUC ranging from 0.747 (support vector machine) to 0.909 (random forest).
To validate the in silico observations, the expression of 29 TMI-comprising genes (obtained from 2 previous studies[7, 8]) in six patient-derived HNSCC lines (NCC-HN1, NCC-HN19, NCC-HN26, NCC-HN43, NCC-HN64, NCC-HN90) relative to a normal squamous epithelial cell line (Het1A) was examined. Using qRT-PCR, 17 genes had detectable mRNA expression in all patient-derived HNSCC cell lines, and they were all differentially expressed in at least one HNSCC line (Figure 1E). In particular, MMP1 and MMP12 upregulation were most robust, with a >10-fold increase in multiple HNSCC lines (Supplementary Table S4). Immunohistochemistry staining of HNSCC tissues including the tongue, floor of the mouth, hypopharynx and larynx confirmed that MMP1 and MMP12 proteins were overexpressed in tumor tissues, compared to paired adjacent normal tissues (Figure 1F, Supplementary Figure S2).
To understand how MMP1 and MMP12 promote tumor progression in HNSCC, MMP1 and MMP12 expression were depleted by siRNA, and the effects on migration, invasion and proliferation were examined. Using in vitro transwell migration, wound healing, matrigel invasion and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays, MMP1 knockdown showed no obvious effects on the migration, invasion and proliferation of NCC-HN26 cells, despite NCC-HN26 exhibiting high endogenous MMP1 expression (Supplementary Figure S3), leading us to hypothesize that MMP1 could be functioning redundantly with another MMPx (possibly other collagenases). In the absence of MMP1, other MMPs that digest similar substrates (e.g., MMP13) or work in the same process could still compensate for the function of MMP1 to promote tumor growth and progression. In contrast, MMP12 knockdown significantly reduced the migration and invasion of NCC-HN26 cells relative to the control (Figure 1G, Supplementary Figure S4A-C), but had no obvious effect on the proliferation of NCC-HN26 (Supplementary Figure S4D). To confirm knockdown efficiency and ensure these observations were not cell-line-specific, we repeated the experiments in NCC-HN1 and NCC-HN19 cells and found that the results were consistent (Supplementary Figure S4E-N). We further performed a double knockdown of MMP1 and MMP12 to test for synergistic effects on tumor cell migration, invasion and proliferation. However, the double knockdown did not cause further reduction in migration, invasion and proliferation of NCC-HN26 cells compared to MMP12 single knockdown (Supplementary Figure S5). Together, these results suggest that MMP12 may promote tumor progression in HNSCC.
We next examined whether MMP1 or MMP12 depletion affected the sensitivity of HNSCC lines to cisplatin and cetuximab. MMP1 knockdown had no obvious effects on response to cisplatin and cetuximab in most HNSCC lines (Supplementary Table S5, Supplementary Figure S6). In contrast, MMP12 knockdown significantly improved the response of the five HNSCC cell lines (NCC-HN26, NCC-HN1, NCC-HN43, NCC-HN64, NCC-HN90) to cetuximab while no obvious effect on cisplatin response was observed (Figure 1H-L, Supplementary Figure S6). To confirm that MMP12 inhibition improves the sensitivity of HNSCC cells to cetuximab, we carried out a combinatorial drug treatment using MMP408 (specific MMP12 inhibitor) at four concentrations (0-0.01mmol/L) and cetuximab at 11 concentrations (0-0.02mmol/L). We found a significant reduction in the IC50 (half-maximal inhibitory concentration) values of cetuximab at higher concentration of MMP408 across several HNSCC lines (Supplementary Table S6, Supplementary Figure S7).
We also examined the prognostic significance of MMP12 expression in the TCGA-HNSC cohort (n = 520). We stratified the patients into two risk groups, MMP12low and MMP12high (Supplementary Figure S8A), using CutoffFinder as previously described [7, 8]. The two groups had significantly different disease-free interval (DFI; HR (Hazard Ratio) = 3.02; log-rank P = 0.058; Supplementary Figure S8B). We further performed multivariate Cox regression survival analyses to adjust for other clinical variables such as clinical stage, targeted treatment, and subsite (Supplementary Table S7-S8). As the multivariate analyses involving all the above-mentioned factors returned infinite coefficients, we performed survival analyses independently adjusting for different sets of covariates and found that MMP12 expression could be a potential independent indicator of prognosis (log-rank P = 0.08 and 0.05, respectively). Of the 520 TCGA-HNSC cases analyzed, there were only 18 cetuximab-treated cases with available DFI survival data, with one patient having experienced the event (i.e. death), resulting in the infinite regression coefficients. Using the disease-specific survival (DSS) endpoint, we found that the probability of survival in MMP12high and MMP12low cetuximab-treated group (cutoff = 8.4) was 0.72 and 0.82 respectively, demonstrating a 10% survival difference between the two risk groups stratified by MMP12 (Supplementary Table S9). We performed Cox regression survival analyses to examine the prognostic performance of HNSC subsite and HPV (Human Papillomavirus) status. Due to limited data, however, it was not possible to observe any significant difference in cetuximab-treated patients (Supplementary Table S10).
Consistent with our current study, a previous report has also implicated MMPs in conferring resistance to cetuximab in HNSCC [10]. Johansson et al. [10] showed that HNSCC cells treated with cancer-associated fibroblasts (CAF) conditioned media promoted resistance to cetuximab treatment. The addition of a general MMP inhibitor to CAF conditioned media partially abolished the resistance to cetuximab in HNSCC cells, indicating that MMPs mediated this resistance [10]. However, the conditioned media collected from CAFs treated with MMP1 siRNAs failed to improve the response of HNSCC to cetuximab, suggesting that other MMPs could be mediating cetuximab resistance [10]. Our results suggest that MMP12 may influence resistance to cetuximab.
In conclusion, MMP12 may promote tumor migration and invasion of HNSCC. Inhibition of MMP12 enhanced the sensitivity of HNSCC cells to cetuximab. These results suggest that MMP12 overexpression may flag more aggressive disease that cannot be improved by cetuximab treatment alone, and that inhibition of MMP12 in addition to the EGFR axis may improve clinical outcomes of HNSCC patients.
DECLARATIONS
ETHICS APPROVAL AND CONSENT
All HNSCC lines were derived from patients who undergo surgery at the National Cancer Centre Singapore in accordance with approval from the SingHealth Centralized Institutional Review Board (CIRB 2007/441/B).
AUTHORS’ CONTRIBUTIONS
DWTL conceived and supervised the study. KYG, SBL, KWL, TYDC, SCT conducted the experiments. KYG, SBL, KWL performed data analysis. KYG wrote the manuscript draft. NGI and CTL contributed intellectual inputs. All authors reviewed and approved the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Elaine Lim for providing helpful discussion. We also thank SingHealth Tissue Repository for providing tissue blocks for staining.
FUNDING
This work was supported by funding from the National Medical Research Council NMRC/CSA-INV/0025/2017, MOH-000375-01 and NCCRF-YR2018-JAN-SUG2. Su Bin Lim is supported by the National Research Foundation of Korea (2021R1F1A1064122, 2020R1A6A1A03043539, and 2020M3A9D8037604) and the new faculty research fund of Ajou University School of Medicine.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no potential conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.




