Interactions among variants in P53 apoptotic pathway genes are associated with neurologic deterioration and functional outcome after acute ischemic stroke
Abstract
Objective
Neurologic deterioration (ND) and functional outcome after ischemic stroke (IS) are not accurately predicted by clinical pictures on admission. The aim of present study was to investigate the association of variants in P53 apoptotic pathway genes with ND and functional outcome after IS.
Methods
Genotypes of nine variants in apoptosis-relevant genes were measured in patients with acute IS. Gene–gene interactions were analyzed by generalized multifactor dimensionality reduction (GMDR). The primary outcome was ND. ND was diagnosed in patients who worsened ≥2 points (National Institutes of Health Stroke Scale [NIHSS] score) within the first 10 days of stroke onset. The secondary outcome was functional status at 90 days after IS as measured by modified Rankin Scale (mRS) score.
Results
A total of 705 enrolled patients, ND occurred in 174 (24.7%) patients, and 184 (26.1%) patients were poor functional outcome (mRS score > 2). Although the nine variants were not significantly associated with ND and functional outcome by univariate analysis, there was a gene–gene interaction among P53rs1042522, MDM-2rs2279744, and MMP-9 rs3918242 using GMDR analysis. The high-risk interaction among the three variants was independently associated with higher risk of ND (HR, 2.04, 95% CI: 1.22–5.64, p = .018) and poor functional outcome (OR, 2.68, 95% CI: 1.68–7.86, p = .004) after adjusting for the covariates.
Conclusion
The interactions among P53 rs1042522, MDM-2 rs2279744, and MMP-9 rs3918242 may increase the risk of ND and poor functional outcome and may be considered as a genetic marker of predicting ND and poor functional outcome after stroke.
1 BACKGROUND
Stroke is the first cause of death and adult disability in China (Guan et al., 2017), with approximately 80% being ischemic strokes (IS). Neurologic deterioration (ND) occurs in one third of patients with acute IS, and it is a devastating complication and associated with increased mortality and long-term functional disability (Vahidy et al., 2014; Yi, Han, Zhou, Lin, & Liu, 2016). However, ND or functional outcome after IS remains largely unpredictable (Weimar, Ziegler, König, & Diener, 2002). Patients with initially a similar clinical picture can worsen or improve dramatically within the first days after IS (Banks & Marotta, 2007; Castillo, 1999). Neuronal apoptosis in the ischemic penumbra may be an important mechanism for ND and impaired functional recovery of IS patients (Broughton, Reutens, & Sobey, 2009; Sairanen, Karjalainen-Lindsberg, Paetau, Ijäs, & Lindsberg, 2006). Thus, the variable prediction of ND or functional outcome after stroke could be the effect of different genetic backgrounds to apoptosis.
Apoptosis is also called programmed cell death, and it is an important mechanism of delayed ischemic brain damage in animal experiments (Sairanen et al., 2006). Two general pathways of apoptosis are triggered after cerebral ischemia, that is, intrinsic pathway (originating from mitochondrial release of cytochrome c) and the extrinsic pathway (originating from the activation of cell surface death receptors) (Broughton et al., 2009). Genetic polymorphisms of cell-cycle regulating genes may affect the DNA damage (Yagnik, Jahangiri, Chen, Wagner, & Aghi, 2017). When cells are damaged, cell division will stop in G1 phase of mitosis (Duan et al., 2017). In the cell-cycle regulation, the arrest of cell cycle in G1/S transform depends on p53 process (Yousefi, Rahmati, & Ahmadi, 2014), which is the master control system of the cell cycle, cell apoptosis, and genome stability. P53 encodes p53 transcription factor, a tumor suppressor protein that can mediate apoptosis in eukaryotic cells. Recent studies have shown P53 Arg72Pro (rs1042522) polymorphism triggers neuronal death via the mitochondrial apoptotic pathway (Gomez-Sanchez et al., 2011). P21, the downstream gene of the P53 gene, can prevent cell-cycle progression in the G1/S and G2/M phases and plays a key role in suppressing cancer (Karimian, Ahmadi, & Yousefi, 2016). The murine double minute 2 (MDM-2) gene is a negative regulator of P53, which can inhibit P53 expression (Moumen, Patane, Porras, Dono, & Maina, 2007), and P53 may upregulate P21 expression in response to DNA damage (Macleod et al., 1995). Rs1042522 polymorphism may affect neuronal vulnerability to apoptosis and can be considered as a genetic marker for poor functional outcome after stroke (Gomez-Sanchez et al., 2011). Many studies have shown that single nucleotide polymorphisms (SNPs) of P53, MDM-2, and P21 rs1801270 play important roles in DNA damage and apoptosis and are intimately related to cancer occurrence (Chen et al., 2015; Duan et al., 2018). Matrix metalloproteinase-9 (MMP-9) can activate numerous pro-inflammatory cytokines and chemokines such as interleukin and tumor necrosis factor, facilitates leukocytes transport across the endothelium, and plays a key role in neuronal damage, apoptosis, and blood–brain barrier (BBB) destruction after cerebral ischemia (Barr et al., 2010; Candelario-Jalil, Yang, & Rosenberg, 2009). Polymorphisms of MMP-9 gene regulate the transcription of MMP-9 protein and are associated with increased IS or cancer risk (Yuan et al., 2013; Zhu, Liu, Zhou, & Chen, 2018). So far, no study has reported the correlations between the SNPs of P53, P21, MDM-2, and MMP-9 genes and ND or functional outcome after IS.
Neurologic deterioration and functional outcome after IS may be very complex, and a single polymorphism in a particular gene is unlikely to explain completely the complex genetic etiology for ND and functional outcome. Gene–gene interactions or gene–environmental interactions may synergistically contribute to ND and functional outcome (Yi, Liao, Fu, Zhang, & Wang, 2015). Generalized multifactor dimensionality reduction (GMDR) analysis is customarily used to assess the higher order gene–gene or gene–environment interactions (Lou et al., 2007). However, the potential effects of gene–gene interactions in apoptotic -relevant genes on ND and functional outcome after IS are unclear. In this study, therefore, we aimed to investigate the association of gene–gene interactions among apoptotic-relevant genes with ND and functional outcome after IS, which could provide more insights into the genetic background for ND and functional outcome, prevent ND for better, and improve functional outcome.
2 METHODS
2.1 Ethics statement
This study was conducted in the People's Hospital of Deyang City and the Third Affiliated Hospital of Wenzhou Medical University between March 2014 and December 2016. The study protocol was approved by the Ethics Committee at the participating hospitals in accordance with the principles stated in the Declaration of Helsinki. Written informed consent was obtained from each of the participants before participating in the study.
2.2 Study population
Between March 2014 and December 2016, we consecutively registered patients who had suffered their first-ever IS and were admitted to the participating hospitals within the first 48 hr after onset of symptoms. IS were confirmed on the basis of both clinical findings and brain magnetic resonance imaging (MRI) scan. All patients underwent computed tomography or MR angiography of the brain, carotid duplex ultrasound, common electrocardiogram (ECG), or 24-hr Holter ECG, as well as echocardiogram. The inclusion criteria were as follows: (a) age ≥ 40 years old and (b) National Institutes of Health Stroke Scale (NIHSS) score ≤ 15 points on admission. Exclusion criteria were as follows: (a) thrombolytic therapy or thrombectomy; (b) NIHSS score > 15 points at admission; (c) severe cardiovascular, liver, and renal disease; (d) other determined etiology or undetermined etiology stroke according to new subtype classification criteria (Han et al., 2007); (e) hypoxia, fever, or any relevant hemodynamic compromise on admission; and (f) unwilling to participate in this study. All enrolled patients received standard therapy according to the guidelines (Jauch et al., 2013; Kernan et al., 2014).
2.3 Clinical variables
Medical history and vascular risk factors were recorded on admission. Fasting blood samples from patients were assessed for glucose, triglycerides (TG), total plasma cholesterol (TC), and low-density lipoprotein cholesterol (LDL-C). Hyperlipidemia was defined as TG > 180 mg/dl, TC > 200 mg/dl, or use of lipid-lowering medication (Yi et al., 2016). Stroke subtypes were classified according to the new subtype classification criteria (Han et al., 2007). Stroke severity was evaluated by a certified member of stroke team using the NIHSS on admission.
2.4 Outcome variables
For each patient, NIHSS was assessed by a member of stroke team on admission and subsequently daily during the period of hospitalization. Functional outcome was evaluated at 3 months using the modified Rankin Scale (mRS). The primary outcome was ND. ND was diagnosed in patients who worsened ≥2 points (NIHSS) within the first 10 days of stroke onset after excluding a new infarct in another vascular territory or hemorrhagic transformation (HT) (Yi et al., 2016). The secondary outcome was functional status at 90 days after IS. mRS score > 2 was considered as poor functional outcome, and mRS score ≤ 2 was defined as good functional outcome (Swieten, Koudstaal, Visser, Schouten, & Gijn, 1988).
2.5 The selection of SNPs and genotyping
In this study, SNPs of apoptotic-relevant genes were selected from the NCBI database (http://www.ncbi.nlm.nih.gov/SNP), according to the following criteria: (a) These SNPs have been evaluated in previous studies (Gomez-Sanchez et al., 2011; Moumen et al., 2007; Yuan et al., 2013; Zhu et al., 2018) and (b) minor allele frequency for these SNPs > 0.05. According to the criteria, nine variants were assessed, including P53 rs1042522, MDM-2 rs2279744, MDM-2 rs1690916, P21 rs1801270, MMP-9 rs1056628, MMP-9 rs3918242, MMP-9 rs17576, MMP-9 rs3787268, and MMP-9 rs2250889.
Genomic DNA from peripheral blood was extracted using a modified phenol/chloroform method and purified using the UNIQ-10 kit (Sangon Biotech Co., Ltd.). The genotyping of these SNPs was performed by authors blinded to the clinical data of patients, using the matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry method, as our previously described (Yi et al., 2015). In brief, each SNP gene possessed a specific genotype, with two amplification primers, and one extension primer. The reaction mix was desalted by adding 6 mg of cation exchange resin (Sequenom Inc.), mixed, and resuspended in 25 µl of water. Once the primer extension reaction was completed, the samples were spotted onto a 384-well spectroCHIP (Sequenom Inc.) using MassARRAY Nanodispenser (Sequenom Inc.) and genotyped using the MALDI-TOF mass spectrometer. Genotype calling was performed in real time with MassARRAY RT software version 3.0.0.4 and analyzed using the MassARRAY Typer software version 3.4 (Sequenom Inc.).
2.6 Statistical analysis
The data were analyzed using SPSS 16.0 software. The results are expressed as percentages for categorical variables, and continuous variables are expressed as mean ± SD. Baseline clinical characteristics and genotype distribution of the nine variants were compared using Student's t test (continuous variables) and chi-square test (categorical variables) between patients with and without ND.
The allele frequencies for Hardy–Weinberg equilibrium were evaluated using chi-square test. The GMDR software was used to assess gene–gene interactions under various scenarios as previously reported (Lou et al., 2007; Yi et al., 2015). In brief, the nine variants were coded from number 1 to 9, and GMDR computes the maximum-likelihood estimates and the scores of all individuals under the null hypothesis. The cumulative score is calculated within each multifactor cell, which is labeled either as high risk if the average score meets or exceeds a pre-assigned threshold of 0 or as low-risk if the score is less than 0. An exhaustive search of all possible one- to ten-locus models was performed for all variants. The model with the minimum prediction error, the maximum cross-validation consistency score, and 0.05 or lower p value derived from the sign test automatically in the GMDR software was considered as the best model, which were confirmed by a permutation test implemented in the GMDR software as well.
Incidence of ND between patients with and without high-risk interactive genotype was compared by chi-square test. The high-risk interaction genotype was assigned as one, and low-risk interaction genotype was assigned as zero in Cox proportional-hazards model and multivariable logistic regression analysis. The independent contribution of gene–gene interaction to ND was assessed using Cox proportional-hazards model after adjusting for covariates (variables with p value < .2 by univariate analysis) and reported as the hazard ratio (HR) with the 95% confidence interval (CI). The influence of the high-risk interactive genotype on functional outcome was investigated by multivariable logistic regression analysis, after adjusting for the main baseline variables related to each main variable in the univariate analysis (enter approach and probability of entry p < .05) and reported as odds ratio (OR) with 95% CI.
All tests were two-sided, and p value < .05 was considered statistically significant.
3 RESULTS
3.1 Clinical characteristics in patients with and without ND
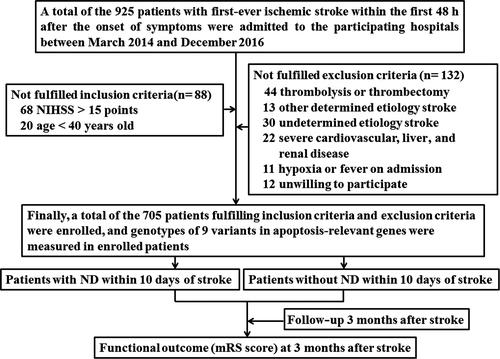
Between March 2014 and December 2016, 925 patients with first-ever IS within the first 48 hr after the onset of symptoms were admitted to the participating hospitals. Among the 925 patients, 220 patients did not fulfill inclusion criteria and exclusion criteria. Finally, a total of the 705 patients fulfilling inclusion criteria and exclusion criteria were enrolled. The detailed procedure in this study was presented in Figure 1. Among the 705 enrolled patients, the duration of in-hospital ranged from 10 to 18 days (median, 13.6 days). There were no patients discharged within 10 days after stroke onset. ND occurred in 174 (24.7%) patients within the first 10 days of stroke onset. Compared with patients without ND, the age was older, and fasting glucose and hemoglobin A1c were higher in patients with ND (Table 1).
| Characteristics |
Patients with ND (n = 174) |
Patients without ND (n = 531) |
p Value |
|---|---|---|---|
| Age (years) | 70.8 ± 14.3 | 68.2 ± 14.7 | .043 |
| Men (n, %) | 100 (57.5) | 295 (55.6) | .673 |
| Hypertension (n, %) | 140 (80.5) | 411 (77.4) | .412 |
| Diabetes mellitus (n, %) | 65 (37.4) | 161 (30.3) | .091 |
| Atrial fibrillation (n, %) | 21 (12.1) | 58 (10.9) | .671 |
| Current smoker (n, %) | 72 (41.4) | 213 (40.1) | .778 |
| Hyperlipidemia (n, %) | 100 (57.5) | 289 (54.4) | .482 |
| Systolic blood pressure (mmHg) | 153.4 ± 14.8 | 152.6 ± 17.5 | .552 |
| Diastolic blood pressure (mm Hg) | 90.1 ± 11.8 | 89.5 ± 15.7 | .593 |
| Fasting glucose (mM) | 7.5 ± 2.7 | 6.6 ± 2.5 | <.001 |
| Hemoglobin A1c (%) | 7.3 ± 2.4 | 6.5 ± 2.2 | <.001 |
| Onset to admission time (h) | 29.9 ± 16.2 | 30.4 ± 17.8 | .731 |
| NIHSS score at admission | 10.9 ± 3.5 | 10.4 ± 3.7 | .125 |
| Stroke subtype (n, %) | |||
| Atherothrombosis | 102 (58.6) | 306 (57.6) | .986 |
| Small artery disease | 41 (23.6) | 134 (25.2) | |
| Cardioembolism | 31 (17.8) | 91 (17.1) | |
| In-hospital treatment (n, %) | |||
| Antihypertensive drugs | 145 (83.3) | 422 (79.5) | .296 |
| Hypoglycemic drugs | 69 (39.7) | 171 (32.2) | .089 |
| Statins | 166 (95.4) | 519 (97.7) | .113 |
| Aspirin | 107 (61.5) | 337 (63.5) | .526 |
| Aspirin plus clopidogrel | 44 (25.3) | 168 (31.6) | .111 |
- Abbreviations: ND, neurologic deterioration; NIHSS, National Institutes of Health Stroke Scale.
3.2 Genotype distributions in patients with and without ND
The genotype distributions of the nine variants were consistent with the Hardy–Weinberg equilibrium (all p > .05). There were no significant differences of genotype distributions in the nine variants between patients with and without ND by univariate analysis (p > .05 for each variant individually, Table 2).
|
Patients with ND (n = 174) |
Patients without ND (n = 531) |
p-Value | |
|---|---|---|---|
| P53 (rs1042522) | |||
| CC | 27 (15.5) | 97 (18.3) | .738 |
| CG | 88 (50.6) | 263 (49.5) | |
| GG | 59 (33.9) | 171 (32.2) | |
| MDM-2 (rs2279744) | |||
| TT | 36 (20.7) | 101 (19.0) | .885 |
| TG | 90 (51.7) | 275 (51.8) | |
| GG | 48 (27.6) | 155 (29.2) | |
| MDM-2 (rs1690916) | |||
| GG | 93 (53.4) | 307 (57.8) | .294 |
| GA | 65 (37.4) | 194 (36.5) | |
| AA | 16 (9.2) | 30 (5.6) | |
| P21 (rs1801270) | |||
| CC | 38 (21.8) | 134 (25.2) | .366 |
| CA | 82 (47.1) | 257 (48.4) | |
| AA | 54 (31.0) | 140 (26.4) | |
| MMP-9 (rs1056628) | |||
| AA | 114 (65.5) | 364 (68.5) | .516 |
| AC | 48 (27.6) | 142 (26.7) | |
| CC | 12 (6.9) | 25 (4.7) | |
| MMP-9 (rs3918242) | |||
| CC | 116 (66.7) | 388 (73.1) | .188 |
| CT | 46 (26.4) | 121 (22.8) | |
| TT | 12 (6.9) | 22 (4.1) | |
| MMP-9 (rs17576) | |||
| AA | 20 (11.5) | 60 (11.3) | .893 |
| AG | 70 (40.2) | 203 (38.2) | |
| GG | 84 (48.3) | 268 (50.5) | |
| MMP-9 (rs3787268) | |||
| AA | 65 (37.4) | 218 (41.1) | .678 |
| AG | 50 (28.7) | 145 (27.3) | |
| GG | 59 (33.9) | 168 (31.6) | |
| MMP-9 (rs2250889) | |||
| CC | 99 (56.9) | 296 (55.7) | .725 |
| CG | 52 (29.9) | 173 (32.6) | |
| GG | 23 (13.2) | 62 (11.7) | |
- Abbreviation: ND, neurologic deterioration.
3.3 Gene–gene interactions
Although the nine variants in apoptotic-relevant genes were not significantly associated with ND by univariate analysis, there was a gene–gene interaction among the nine variants using GMDR analysis. The best model for ND was interaction among P53 rs1042522, MDM-2 rs2279744, and MMP-9rs3918242 after adjusting for confounding variables (p = .021, Table 3). The one-locus model was computed for each variant and the empirical p values for prediction error using permutation testing were .026, indicating the interactions among the three variants synergistically contributed to a higher risk of ND than did single variant alone.
| Best model6 | Training balanced accuracy | Testing balanced accuracy | Cross-validation consistency | Sign test (p value) |
|---|---|---|---|---|
| 1 | 0.523 | 0.486 | 7/10 | 8 (.421) |
| 1,2 | 0.581 | 0.523 | 8/10 | 7 (.227) |
| 1,2,3 | 0.632 | 0.597 | 10/10 | 9 (.021) |
| 1,2,4,5 | 0.504 | 0.468 | 9/10 | 6 (.137) |
| 1,2,3,4,5 | 0.584 | 0.467 | 6/10 | 8 (.359) |
| 1,2,3,4,5,6 | 0.611 | 0.543 | 5/10 | 6 (.554) |
| 1,2,3,4,5,6,7 | 0.425 | 0.611 | 7/10 | 6 (.472) |
| 1,2,3,4,5,6,7,8 | 0.542 | 0.493 | 9/10 | 6 (.535) |
| 1,2,3,4,5,6,7,8,9 | 0.798 | 0.522 | 8/10 | 6 (.633) |
- Abbreviations: GMDR, generalized multifactor dimensionality reduction; ND, neurologic deterioration.
- Numbers 1–9 represent rs1042522, rs2279744, rs3918242, rs1690916, rs1801270, rs1056628, rs3787268, rs17576, and rs2250889, respectively.
3.4 Associations between different genotype combinations and ND risk
Then, we evaluated the relationship of different genotype combinations of the three interactive variants with the risk of ND. The wild-type genotype for the three variants was used as the reference. Compared to the patients harboring wild-type genotype rs1042522CC, rs2279744TT, and rs3918242CC, the relative risk of different genotype combinations among the three variants for ND was assessed. The risks for ND were higher in patients harboring rs1042522GG, rs2279744GG, and rs3918242TT; rs1042522GG, rs2279744GG, and rs3918242TT/CT; and rs1042522GG, rs2279744TG, and rs3918242CT, compared with those carrying rs1042522CC, rs2279744TT, and rs3918242CC (Table 4). The three combination genotypes of rs1042522, rs2279744, and rs3918242 were defined as high-risk interactive genotype. The other combination genotypes of rs1042522, rs2279744, and rs3918242 did not reach statistical significance level of 0.05 (Table 4) and were considered as low-risk interactive genotype.
| Rs1042522 | CC | GG | GG | GG | CG | GG, CG | CG | GG, CG |
|---|---|---|---|---|---|---|---|---|
| Rs2279744 | TT | GG | GG | TG | TG | GG | GG, TG | GG, TG |
| Rs3918242 | CC | TT | TT, CT | CT | CT | TT | TT | TT, CT |
| OR | 18 | 2.76 | 2.12 | 1.89 | 1.52 | 1.24 | 1.13 | 1.08 |
| 95% CI | — | 1.31–5.96 | 1.18–5.16 | 1.06–4.58 | 0.97–2.36 | 0.95–2.88 | 0.86–2.05 | 0.83–1.95 |
| p Value | — | .003 | .008 | .027 | .198 | .387 | .497 | .586 |
- Abbreviations: CI, confidence interval; ND, neurologic deterioration; OR, odds ratio.
- The wild-type genotype for each variant was used as the reference.
3.5 Association of high-risk interactive genotype with risk of ND
The incidence of ND was significantly higher in patients carrying high-risk interactive genotype than those carrying low-risk interactive genotype (33.2% [65/196] vs. 21.4% [109/509], p < .001). The risk for ND conferred by high-risk interactive genotype was evaluated using Cox proportional-hazards model. The high-risk interaction was assigned as one, and low-risk interaction was assigned as zero. The other predictors with p value < .2 by univariate analysis also entered in Cox proportional-hazards model for ND, including age, diabetes mellitus, fasting blood glucose, hemoglobin A1c, NIHSS score at admission, statins, aspirin plus clopidogrel, and rs3918242CT/TT. The results showed the high-risk interaction among rs1042522, rs2279744, and rs3918242 was independently associated with higher risk of ND after adjusting for the covariates (HR, 2.04, 95% CI: 1.22–5.64, p = .018, Table 5).
| Factor | HR | 95% CI | p Value |
|---|---|---|---|
| Age | 0.85 | 0.72–1.46 | .453 |
| Diabetes mellitus | 0.91 | 0.85–1.95 | .322 |
| Fasting blood glucose | 1.73 | 1.02–2.76 | .041 |
| Hemoglobin A1c | 1.34 | 0.96–1.86 | .106 |
| NIHSS score at admission | 1.21 | 0.92–2.13 | .412 |
| Statins | 0.86 | 0.68–1.22 | .633 |
| Aspirin plus clopidogrel | 0.72 | 0.53–0.99 | .033 |
| Rs3918242CT/TT | 1.36 | 0.92–2.36 | .286 |
| High-risk interactive variable | 2.04 | 1.22–5.64 | .018 |
Note
- HR for continuous variables means per 1 − SD increase.
- Abbreviations: CI, confidence interval; HR, hazard ratio; ND, neurologic deterioration.
3.6 Association of high-risk interactive genotype with poor functional outcome
Using the mRS to evaluate the disability or dependence in daily living activities of stroke victims at 3 months, we found 184 (26.1%) patients were poor functional outcome (mRS score > 2). There were no significant differences in poor functional outcome among genotypes of the 9 variants. However, the percentage of poor functional outcome was significantly higher in patients carrying the high-risk interactive genotype than those carrying the low-risk interactive genotype (35.6% [84/236] vs. 21.3% [100/469], p < .001). After adjustment for the covariates, including age, diabetes mellitus, hypertension, fasting blood glucose, NIHSS score at admission, neurologic deterioration, and aspirin plus clopidogrel, the high-risk interaction among rs1042522, rs2279744, and rs3918242 was an independent predicting marker of poor functional outcome (OR, 2.68, 95% CI: 1.68–7.86, p = .004, Table 6), as revealed by multivariate logistic regression analysis.
| Factor | OR | 95% CI | p Value |
|---|---|---|---|
| Age | 0.87 | 0.68–1.42 | .637 |
| Diabetes mellitus | 0.86 | 0.72–1.17 | .623 |
| Hypertension | 1.01 | 0.92–1.85 | .564 |
| NIHSS scores on admission | 1.82 | 1.06–3.46 | .024 |
| Neurologic deterioration | 3.68 | 1.94–9.68 | .002 |
| Fasting blood glucose | 1.04 | 0.94–2.35 | .352 |
| Aspirin plus clopidogrel | 0.86 | 0.67–1.02 | .132 |
| High-risk interactive variable | 2.68 | 1.68–7.86 | .004 |
Note
- OR for continuous variables means per 1 − SD increase.
- Abbreviations: CI, confidence interval; mRS, modified Rankin Scale; NIHSS, National Institutes of Health Stroke Scale; OR, odds ratio.
4 DISCUSSION
In this study, we found that 174 (24.7%) patients suffered from ND, and 184 (26.1%) patients were poor functional outcome (mRS score > 2). Although the nine variants in apoptotic-relevant genes were not associated with ND and functional outcome by univariate analysis, GMDR analysis revealed that there was a gene–gene interaction among P53 rs1042522, MDM-2 rs2279744, and MMP-9 rs3918242, and the high-risk interaction among the three variants was independently associated with higher risk of ND and poor functional outcome.
Some studies have revealed that low circulating levels of retinoic acid and plasma neuroendocrine biomarkers, including baseline plasma brain natriuretic peptide, N-terminal pro-brain natriuretic peptide, and cortisol and copeptin levels on admission can predict outcomes and mortality after acute IS (Tu, Dong, Zhao, Yang, & Chen, 2013; Tu et al., 2019). Neuronal apoptosis is an important mechanism of delayed ischemic brain damage in animal experiments (Sairanen et al., 2006). The presence of apoptotic neurons in the ischemic penumbra is associated with poor functional prognosis and mortality after acute IS (Gomez-Sanchez et al., 2011). p53-mediated neuronal death plays a central role of stroke pathophysiology in a mouse model of focal permanent cerebral ischemia (Liu et al., 2013). However, the possible role of SNPs of p53 apoptotic pathway relevant genes in ND and functional outcome after IS has not been thoroughly understood.
The detailed information of the 9 SNPs in this study was summarized in Table 7. Extensive evidences have shown that SNPs of P53 and MDM2 rs2279744 play important roles in DNA damage and cell apoptosis and were independently related to high risk of cancer (Chen et al., 2015; Duan et al., 2018; Liu et al., 2011). Human P53 Arg72Pro (rs1042522) SNP controls susceptibility to ischemia-induced neuronal apoptosis and the functional outcome after stroke (Gomez-Sanchez et al., 2011). P53 rs1042522 SNP can condition neuronal ischemic tolerance by modulating mitochondrial p53 stabilization (Ramos-Araque et al., 2018). MMP-9 polymorphisms regulate the transcription of MMP-9 protein and are associated with increased IS or cancer risk and hemorrhagic transformation of IS (Yuan et al., 2013; Zhang, Cao, Xu, Li, & Xu, 2015; Zhu et al., 2018). In this study, we did not find the association of nine variants in apoptotic-relevant genes with ND and functional outcome by univariate analysis. However, the most noteworthy finding in this study was that there was a gene–gene interaction among rs1042522, rs2279744, and rs3918242 using GMDR analysis, and the high-risk interaction among the three variants was independently associated with risk of ND and poor functional outcome. This indicates interaction among the three variants synergistically contributes to a higher risk of ND and poor functional outcome than do single variant alone.
| Chromosomal location | Types of amino acid changes | Functional consequence | Results of studies or clinical significance: | |
|---|---|---|---|---|
| P53(rs1042522) | 17:7676154 | Pro72Arg | Missense variant | Uncertain-significance or drug-response |
| MDM-2(rs2279744) | 12:68808800 | N/A | Intron variant | Accelerated tumor formation and risk |
| MDM-2(rs1690916) | 12:68841626 | N/A | 3 Prime UTR variant | Not Reported in Clin |
| P21(rs1801270) | 6:36684194 | Ser31Arg | Missense variant | Tumor risk |
| MMP-9(rs1056628) | 20:46016407 | N/A | Intron variant and 3 Prime UTR variant | Not Reported in Clin |
| MMP-9(rs3918242) | 20:46007337 | N/A | 2KB upstream variant | Not reported in Clin |
| MMP-9(rs17576) | 20:46011586 | Gln279Arg | Missense variant | Metaphyseal anadysplasia |
| MMP-9(rs3787268) | 20:46013092 | N/A | Intron variant | Not Reported in Clin |
| MMP-9(rs2250889) | 20:46013767 | Arg574Leu | Missense variant | Metaphyseal anadysplasia |
- Abbreviation: SNPs, single nucleotide polymorphisms.
The nature of the gene–gene interaction among the three variants is not clear. It may be due to rs1042522, rs2279744 and rs3918242 may synergistically affect cell-cycle regulation, DNA damage, cell apoptosis, and BBB destruction (Chen et al., 2015; Duan et al., 2018; Gomez-Sanchez et al., 2011; Yousefi et al., 2014), which contribute to a higher risk of ND and poor functional outcome after IS. In previous reports, P53 encodes p53 tumor suppressor protein that can mediate apoptosis in eukaryotic cells (Yousefi et al., 2014), and it is a tumor suppressor gene involved in the G1-S checkpoint and has the function of gene guarding (Pietsch, Humbey, & Murphy, 2006). P53 rs1042522 SNP occurs in a proline-rich domain involved in the proapoptotic function of p53 (Pietsch et al., 2006; Sakamuro, Sabbatini, White, & Prendergast, 1997). Variant of P53 rs1042522 is a potent inducer of apoptosis and inhibitor of oncogenic transformation and determines the age of onset and cancer progression (Pietsch et al., 2006; Whibley, Pharoah, & Hollstein, 2009; Zhu et al., 2010). Gomez-Sanchez et al. (2011) reported P53 Arg/Arg genotype was linked to early ND and poor functional outcome after IS. In primary cultured neurons, Arg72-p53 interacted directly with mitochondrial B-cell lymphoma-extra large activated the intrinsic apoptotic pathway and ischemia-induced apoptotic cell death. Delayed treatment with a p53 inhibitor (pifithrin-alpha) may modify stroke-induced endogenous neurogenesis and improve the functional recovery in stroke animals (Luo et al., 2009). MDM-2 is an important regulator of P53 and has the function of degrading P53 (Qiu et al., 2008). Although there was no report regarding the relationship between MDM-2 genetic polymorphisms and ND or functional outcome after IS, some studies have shown that MDM-2 polymorphisms may be a risk factor for uterine fibroids and hepatocellular carcinoma (Dong et al., 2012; Salimi et al., 2015). This was in good agreement with the highlighted notion that neuronal death and oncogenesis may share common mechanistic foundations (Morris, Veeriah, & Chan, 2010). MMP-9 can activate numerous pro-inflammatory cytokines and chemokines and involves in neuronal damage and apoptosis (Barr et al., 2010; Candelario-Jalil et al., 2009). Previous studies showed that MMP-9 polymorphisms were associated with increased risk of IS, cancer, and hemorrhagic transformation of IS (Yuan et al., 2013; Zhang et al., 2015; Zhu et al., 2018). Up to date, there were no studies to investigate the association of interaction among the SNPs of P53, P21, MDM-2, and MMP-9 with ND or functional outcome after IS. However, Duan et al. (2018) has revealed that interaction among P53, P21, and MDM-2 may affect cholinesterase activity in people with exposure to omethoate. Thus, we reason that the high-risk interaction among P53, MDM-2, and MMP-9 could synergistically activate the intrinsic apoptotic pathway and ischemia-induced apoptotic cell death, thereby increasing the risk of ND and poor functional outcome after IS.
Despite our findings are interesting, there are several limitations in this study. First, this study was performed in two-centers, and the samples were small. Thus, our findings should be validated in larger samples, multicenter studies. Second, although we genotyped known functional variants in P53, P21, MDM-2, and MMP-9 gene, some rare functional variants were not assessed in this population. Furthermore, apoptotic pathways are very complex, and many genes may involve in regulating apoptosis. In this study, we only investigated the association of nine variants in P53 apoptotic pathway with ND and functional outcome. Thus, future studies involving a larger set of genetic variants must be conducted to investigate the full extent of gene–gene interaction effect on ND and functional outcome. Third, although we found the three variants in P53, MDM-2, and MMP-9 could synergistically contribute to a higher risk of ND and poor functional outcome, we did not investigate the molecular mechanisms of the gene–gene interactions. Therefore, in the next study we will plan to use the primary cultured neurons or animal models of cerebral ischemia to explain the molecular mechanisms of interaction among the three variants. Finally, lack of an independent sample for replication was also a limitation in this study.
5 CONCLUSION
The incidence of ND and poor functional outcome after stroke is very common. There is a gene–gene interaction among P53 rs1042522, MDM-2 rs2279744, and MMP-9 rs3918242. The high-risk interaction among the three variants may increase the risk of ND and poor functional outcome and may be considered as a genetic marker of predicting ND and poor functional outcome after IS.
ACKNOWLEDGMENTS
This study was supported in part by grants from the Sichuan Science and Technology Agency Research Foundation (Grant No.2018JY0164) and the Scientific Research Foundation of Sichuan Provincial Health Department (Grant No. 16ZD046).
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




