Integrative analysis of genome-wide association study and transcriptomics to identify potential candidate genes influencing drip loss in Beijing Black pigs
Jingjing Tian as a co-first author.
Abstract
Understanding the genetic factors related to meat drip loss is of great importance for animal breeding and production. In this study, we employed a combination of genome-wide association study (GWAS) mapping and RNA sequencing (RNA-seq) data to effectively identify potentially functional single nucleotide polymorphisms (SNPs) as well as candidate genes associated with drip loss (DL) in Beijing Black pigs. Initially, we conducted a single- and multi-trait GWAS on drip loss traits in 441 Beijing Black pigs at 24 (DL24) and 48 (DL48) hours postmortem using the Illumina pig 50K SNP chip. Five SNPs with annotations for four genes (FGGY, LHFPL6, OSBPL1A, and NMNAT3) were consistently identified in single or multiple trait GWAS results, indicating their potential pleiotropic effects on drip loss. Next, a comprehensive comparative transcriptomic analysis was performed on samples of Beijing Black pigs exhibiting extremely high and low drip loss, resulting in the identification of 21 differentially expressed genes (DGEs) as potential candidates. Additionally, protein–protein interaction (PPI) network analysis revealed reciprocal regulatory relationships between FOXO1, OSBPL1A, DOCK1 (identified from GWAS) and the candidate DGEs obtained from RNA-seq data. Therefore, we propose that these genes may impact drip loss traits through gene interactions. In conclusion, our integrative analysis screened candidate genes that may affect the drip loss traits in Beijing Black pigs, which provides crucial insights into the molecular mechanisms of drip loss and serves as a theoretical reference for improving meat quality in Beijing Black pigs.
INTRODUCTION
Water holding capacity (WHC) is a crucial parameter for assessing meat quality as it pertains to the ability of meat to retain its inherent water content through a series of physiological and biochemical reactions during postmortem storage [1]. WHC is typically assessed through the measurement of drip loss (DL), which involves the natural exudation of water from meat under gravity at a temperature of 4°C, resulting in subsequent weight reduction [2]. As a result, it is widely acknowledged that meat with lower water holding capacity tends to exhibit higher drip loss, which can directly impact the sensory and visual qualities of the final product [3]. Therefore, enhancing water holding capacity remains a significant challenge for both pork production and meat processing industries.
Beijing Black pig is highly favored by consumers due to its superior meat quality and flavor. This breed is a hybrid cross between Chinese native pig breeds (Dingxian, Shenxian, and Zhouxian) and lineage from Berkshire and Large White [4]. Due to the inter-population specificity, there may be genetic segregation within the Beijing Black pig population, which could potentially impact phenotypic variation in drip loss. Drip loss is a complex trait with heritability estimates ranging from 0.01 to 0.31 [5]. The considerable variability observed in meat drip loss can be attributed to a combination of multigenetic and complex environmental factors [6]. Furthermore, genetic modifications affecting muscle fiber structure and metabolic regulation may play a crucial role in explaining the wide-ranging diversity [7, 8].
Genome-wide association study (GWAS) has emerged as an effective approach for exploring the correlation between genetic variants and phenotypic traits in both humans and farm animals. In this regard, several previous studies have focused on meat quality traits using single-trait models [9]. However, the pleiotropy of genes and genetic correlations between biological traits often lead to false positive results in GWAS when single trait analysis is employed as it fails to account for the correlation among multiple traits [10]. In comparison to single-trait analysis in GWAS, multi-trait model analysis can effectively integrate genetic correlation information among traits, thereby enhancing statistical power efficiency and accuracy [11]. Therefore, a multi-trait GWAS analysis was conducted in this study to identify significant SNPs and candidate genes for drip loss at different postmortem time points (24 and 48 h) in Beijing Black pigs.
Recently, gene expression plays a crucial role in the transition from genotype to phenotype. RNA sequencing (RNA-seq) profiling has been demonstrated as an effective and comprehensive strategy for elucidating significant phenotypic differences by acquiring nearly all transcriptional sequence information of a species at a certain stage [12]. Furthermore, it has been verified that the SNPs loci linked to traits identified by GWAS could result from regulatory mutations within genes instead of amino acid substitutions. As a result, the nearest gene to these SNPs is frequently not responsible for causing the trait [13]. Therefore, the integration of RNA-seq and GWAS analysis not only mutually validates their results but also narrows down the scope to key genes, achieving accurate mapping of complex traits from SNPs to gene expression. This approach enables the identification of mutations affecting mRNA expression at the DNA level and provides more biological explanations for GWAS findings [14].
In this study, we integrated GWAS mapping and RNA-seq data to effectively identify potentially functional SNPs as well as candidate genes associated with drip loss in Beijing Black pigs. First, GWAS was conducted to identify SNPs associated with the drip loss trait in Beijing Black pigs. Second, RNA-Seq was utilized to detect the differentially expressed genes (DEGs) exhibiting distinct expression patterns between high and low drip loss groups. Third, candidate SNPs associated with DEGs were selected for further evaluation of their potential functional relevance to the drip loss phenotype of Beijing Black pigs.
MATERIALS AND METHODS
Sample collection
In accordance with the “Guide for the Care and Use of Laboratory Animals” published by the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China), Beijing Black pigs (Beijing Heiliu Husbandry Technology Co., Ltd.) were humanely sacrificed via electric shock at the Beijing Fifth Meat Processing Factory. The experimental protocols employed in this study were approved by the Animal Care and Use Committee (No. IASCAAS-PG-39).
A total of 441 longissimus dorsi samples were collected from the reciprocal 3-4 rib carcass of Beijing Black pigs (263 males and 178 females) slaughtered at an age range of 210 ± 40 days. The management of all pigs adheres to the protocols established by Beijing Heiliu Husbandry Technology Co., Ltd., ensuring uniform feed composition and feeding environment for all animals. The samples were marked with ear numbers and utilized for determining muscle drip loss values.
Phenotype measurements
Drip loss was determined using the bag method, as previously reported by Otto (2004) [2], whereby approximately 120 g of muscle samples were suspended in an airtight container via a hook under the lid and stored at 4°C. After 24 h (DL24) and 48 h (DL48) of storage, the meat slices were weighed to determine changes in sample weight expressed as a percentage. The estimation of heritability and genotypic correlation was conducted using the GCTA (Genome-wide Complex Trait Analysis) software, while the phenotypic correlation coefficient was measured through Pearson correlation analysis in R packages.
Genotyping and quality control
Genomic DNA was extracted using the salting-out method, and genotyping was performed with the Illumina Porcine 50K BeadChip (Illumina). Quality control procedures were implemented using PLINK software (v.1.90) according to the following criteria: SNPs loci with a call rate less than 95% and a minor allele frequency lower than 0.05 were excluded as well as individuals with a call rate less than 90%. After undergoing quality control, a total of 35,436 SNPs were utilized for subsequent GWAS analyses.
Single- and multi-trait GWAS analyses
The Bonferroni correction method was utilized to determine the threshold values. The p-values for genome-wide significance and suggestive were set at −log10 (0.05 × N/SNPs), where N represents the number of SNPs with a p-value less than 0.05. Finally, R package qqman was employed to generate a Manhattan plot and a quantile–quantile (Q-Q) plot [16].
Annotation of candidate genes
The BioMart bioinformatics database (http://www.ensembl.org/, accessed on 27 August 2022) was utilized to identify candidate genes located within significant and suggestive SNP loci. Specifically, candidate genes situated in the ±50 kb region surrounding SNPs were included in the analysis.
Transcriptome analysis
Based on the drip loss phenotype of all samples, we selected 9 samples each from the highest or lowest drip loss phenotype groups at both 24 and 48 h. After excluding an outlier from the low drip loss group, we ultimately chose 9 samples with high drip loss (H) and 8 samples with low drip loss (L) for RNA-seq analysis. Briefly, RNA samples were isolated using TRIzol reagent (Invitrogen) and library construction was performed using a TruSeq RNA Sample Prep kit (Illumina). Paired-end sequencing was constructed using the Illumina HiSeq 2500 platform (Illumina), and the filtered sequence reads were realigned to the Sscrofa 11.1 reference genome from the Ensembl annotation system. The RNA-seq bioinformatics analysis was conducted following the previously described protocol [17]. The sequencing data used in this study are available at the Genome Sequence Archive, BioProject: CRA011417 (https://ngdc.cncb.ac.cn/).
Statistical analysis
The measurement results of meat drip loss traits were evaluated using the independent-sample t-test procedure and all data presented in the table are reported as means ± standard deviation (Mean ± SD).
RESULTS
Phenotypic characterization of drip loss in Beijing Black pigs
The phenotypic statistics of drip loss in 441 Beijing Black pigs' longissimus dorsi muscles are presented in Table 1. The mean values for DL24 and DL48 were determined to be 0.61% and 1.48%, respectively, indicating that approximately 59% of the drips occurred within the first 24–48 h post-sampling. Furthermore, a high coefficient of variation in drip loss was observed at each time point with a greater value of 149% observed for DL24. Additionally, all phenotypic data underwent Kolmogorov–Smirnov testing for normality as illustrated in Figure S1.
| Traits | Number | Mean ± SDa | Minimum | Maximum | CVb (%) | Heritability (h2) |
|---|---|---|---|---|---|---|
| Drip loss after 24 h storage (DL24), % | 441 | 0.61 ± 0.91 | 0.01 | 9.85 | 149.57 | 0.094 |
| Drip loss after 48 h storage (DL48), % | 441 | 1.48 ± 1.36 | 0.13 | 11.75 | 92.04 | 0.253 |
- a Standard deviation.
- b Coefficient of variation.
Further analysis revealed that the estimated genomic heritability values for DL24 and DL48 were 0.09 and 0.25, respectively (Table 1). This confirms that drip loss-related genetic characteristics are low to medium heritability traits in Beijing Black pigs. Moreover, the phenotypic and genotypic correlations between DL24 and DL48 traits were found to be 0.756 and 0.872, respectively, indicating a strong positive correlation in both aspects.
Population structure in Beijing Black pigs
The population structure of Beijing Black pigs was assessed through principal component analysis (PCA) based on Illumina Porcine 50K BeadChip data as illustrated in Figure 1. To account for population stratification, the first two principal components were incorporated as covariates in the GWAS analysis model.
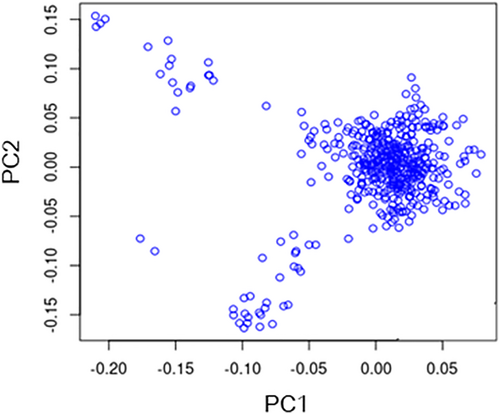
The population structure diagram of Beijing Black pigs.
Summary of the single-trait GWAS
Following rigorous quality control measures, a comprehensive set of 35,436 SNPs was extracted from the porcine assembly genome (Sscrofa 11.1) to facilitate GWAS analysis of drip loss traits in a cohort of 441 Beijing Black pigs. Initially, single-trait GWAS analyses were performed for DL24 and DL48 traits. As showed in Manhattan plots (Figure 2A,B), 8 SNPs loci associated with the DL24 trait that exceeded the chromosomal significance level were identified along with 7 annotated genes nearest to these SNPs. Furthermore, 3 SNPs loci were found to be significantly associated with the DL48 trait, and 5 annotated genes were discovered. The detail information of these SNPs loci and annotated genes information are presented in Table 2.
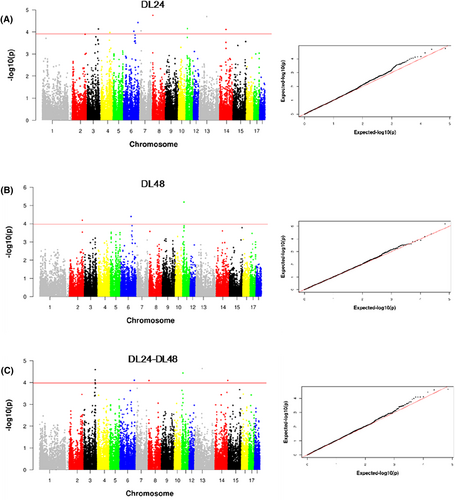
Single-trait and multi-trait GWAS for the drip loss traits (A: DL24, B: DL48, C: DL24-DL48) in Beijing Black pigs. For the Manhattan plots, the y axis represents the negative log10 of p-values for SNPs, while the x axis shows their genomic positions on different chromosomes. The red solid lines indicate suggestive thresholds.
| Trait | SSC | SNP name | SNP position (bp) | p-value | Candidate gene |
|---|---|---|---|---|---|
| DL24 | 3 | WU_10.2_3_127106026 | 119,031,289 | 7.25 × 10−5 | - |
| 6 | WU_10.2_6_140385000 | 152,858,710 | 3.74 × 10−5 | FGGY | |
| 6 | BGIS0007213 | 109,061,542 | 9.35 × 10−5 | OSBPL1A | |
| 7 | 7_13861618 | 13,016,154 | 8.96 × 10−5 | STMND1 | |
| 8 | WU_10.2_8_6359178 | 6,977,551 | 1.76 × 10−5 | - | |
| 11 | INRA0035471 | 15,407,943 | 7.00 × 10−5 | FOXO1, MRPS31 | |
| 13 | CASI0007872 | 80,594,858 | 2.01 × 10−5 | NMNAT3 | |
| 14 | H3GA0040737 | 65,213,534 | 7.63 × 10−5 | CABCOCO1 | |
| DL48 | 2 | M1GA0003187 | 136,836,597 | 6.49 × 10−5 | JADE2, SAR1B, SEC24A |
| 6 | BGIS0007213 | 109,061,542 | 4.04 × 10−5 | OSBPL1A | |
| 11 | ASGA0049925 | 14,449,177 | 6.46 × 10−5 | LHFPL6 | |
| DL24–DL48 | 3 | WU_10.2_3_127106026 | 119,031,289 | 2.57 × 10−5 | - |
| 3 | WU_10.2_3_126377494 | 118,347,227 | 7.79 × 10−5 | - | |
| 6 | WU_10.2_6_140385000 | 152,858,710 | 7.94 × 10−5 | FGGY | |
| 8 | WU_10.2_8_6359178 | 6,977,551 | 8.17 × 10−5 | - | |
| 11 | ASGA0049925 | 14,449,177 | 3.63 × 10−5 | LHFPL6 | |
| 13 | CASI0007872 | 80,594,858 | 2.28 × 10−5 | NMNAT3 | |
| 14 | ALGA0083077 | 136,735,013 | 8.07 × 10−5 | DOCK1 |
- Abbreviation: SSC, sus scrofa chromosome.
Through single-trait GWAS, a total of 11 SNPs loci and 12 annotated genes significantly associated with drip loss in Beijing Black pigs were identified. Among them, only OSBPL1A (SSC6: BGIS0007213) was consistently detected in both DL24 and DL48 single-trait GWAS datasets.
Summary of the multi-trait GWAS
Given the strong genotypic correlation between DL24 and DL48 traits, a multi-trait GWAS was conducted to assess their interaction effects. The result demonstrates that 7 significant SNPs loci and 4 annotated genes were associated with the combined DL24-DL48 trait (Figure 2C). Furthermore, in combination with the findings of single-trait GWAS analysis, FGGY (SSC6: WU_10.2_6_140385000), LHFPL6 (SSC11: ASGA0049925), and NMNAT3 (SSC13: CASI0007872) were repeatedly detected in both single-trait and multi-trait GWAS, indicating their potential role as pleiotropic genes influencing dripping traits and warranting further investigation (Table 2).
Transcriptomic analysis of longissimus dorsi between high and low drip loss groups in Beijing Black pigs
Significant variations in drip loss phenotypes were observed among the Beijing Black pig population, resulting in the classification of samples into high (H) and low (L) groups based on their respective drip loss values. Subsequently, RNA-seq comparison was conducted to investigate potential mechanisms and elucidate detailed phenotypic characteristics. Supplementary Table S1 and Figure S2 provide information for the phenotypic profiles of the samples. A PCA score plot (Figure 3A) demonstrated a distinct separation trend between H and L groups, with a total of 880 differentially expressed genes (DEGs) identified (P ≤ 0.05; Fold Change ≥2 or ≤0.5), including 531 upregulated and 349 downregulated DEGs in the H group compared to the L group (Figure 3B, Table S2).
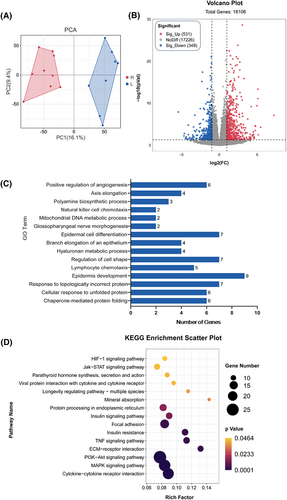
Transcriptomic analysis of longissimus dorsi between high (H) and low (L) drip loss groups in Beijing Black pigs. (A) Principal component analysis (PCA) scores plot derived from longissimus dorsi transcriptomics between the high (H) and low (L) drip loss groups in Beijing Black pigs. (B) Volcano plot of DEGs between the H and L groups in Beijing Black pigs. (C) The top 15 GO terms significantly enriched between the H and L groups in Beijing Black pigs. (D) The top 15 KEGG pathways significantly enriched between the H and L groups in Beijing Black pigs.
Subsequently, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses for the DEGs between the H group and L group. The top 15 significantly enriched GO terms (P < 0.05) were screened out and illustrated in Figure 3C. The DEGs were predominantly enriched in GO terms related to protein folding, epidermal cell development, natural killer cell chemotaxis, regulation of angiogenesis, and mitochondrial DNA metabolic processes. The majority of the biological processes were associated with signal transduction, cellular processes, and myogenesis. Furthermore, Figure 3D displays the top 15 pathways significantly enriched with respect to the drip loss trait, including cytokine–cytokine receptor interaction, MAPK signaling pathway, PI3K-Akt signaling pathway, ECM–receptor interaction, TNF signaling pathway, focal adhesion, protein processing in endoplasmic reticulum, and so on. This suggests that these pathways may play crucial roles in the regulation of drip loss.
Moreover, a considerable number of DEGs were found to be involved in multiple KEGG pathways among the top enriched pathways. Therefore, potential candidate genes were further screened from DEGs that were enriched in more than three KEGG pathways. In the end, a total of 21 genes, including ITGA5, ITGA6, TNC, HSPA1L, HSPA6, and others, were identified (Figure 4A). The information of physiological function on these genes is provided in Table 3.
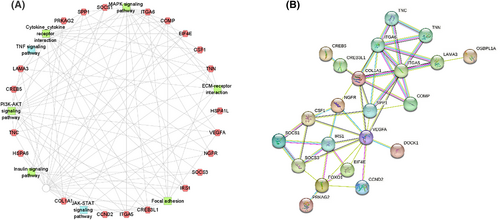
Identification of key candidate genes associated with the drip loss trait in Beijing Black pigs. (A) Network diagram of the candidate DEGs and their corresponding KEGG pathways influencing the drip loss trait in Beijing Black pigs. (B) Protein–protein interaction (PPI) network for the relationship between GWAS-derived genes and RNA-seq-based candidate DEGs.
| Physiological function | Gene name |
|---|---|
| Extracellular matrix protein regulation and integrin binding activity | ITGA5, ITGA6, TNC, TNN, LAMA3, COL1A1, COMP, SPP1 |
| Cytokine signaling regulation | CSF1, SOCS1, SOCS3 |
| Response to insulin signaling | IRS1, NGFR |
| Response to stress | HSPA1L, HSPA6 |
| Transcription factors with high affinity for cAMP-response elements | CREB3L1, CREB5 |
| Fatty acid and cholesterol biosynthesis regulation | PRKAG2 |
| Angiogenesis | VEGFA |
| Cell cycle regulator | CCND2 |
| Eukaryotic translation initiation | EIF4E |
Comprehensive screening of key candidate genes
Based on the GWAS results for DL24 and DL48 as well as RNA-seq analysis between high and low drip loss groups, we investigated whether the annotated genes identified by GWAS were also differentially expressed in RNA-seq results. However, no coexisting genes were identified upon the integration of GWAS and RNA sequencing results. Nevertheless, candidate genes derived from GWAS and RNA-seq data were underwent protein–protein interaction (PPI) network analysis to elucidate the interplay among these putative genetic targets. The PPI network analysis revealed a strong correlation between FOXO1, OSBPL1A, and DOCK1 genes from the GWAS results and the candidate DGEs in RNA-seq data (Figure 4B), highlighting their significant role in the network regulation of drip loss in Beijing Black pigs.
DISCUSSION
The quantity and distribution of water within meat significantly impact its quality with substantial fluid losses in the form of drip potentially compromising pork's financial output, nutritional value, consumer appeal, and technological characteristics [18]. Therefore, a more comprehensive understanding of drip loss traits would prove advantageous for enhancing the economic value of pork. In recent years, the Beijing Black pig has made a significant contribution to the high-quality pork market due to its exceptional meat quality and flavor. Thus, this study represents the first systematic investigation into the breed contribution on meat drip loss traits in Beijing Black pigs through an integrated analysis of GWAS and RNA-seq.
GWAS has been widely employed as an effective approach to directly associate complex traits with genome-wide SNPs for the genetic markers selection, which may impact important economic traits in both animals and plants [19]. Based on genotyping with the Illumina Porcine 50K BeadChip microarray, this study conducted a GWAS analysis of drip loss traits associated with meat quality in Beijing Black pigs. At present, most GWAS studies associate a single trait's phenotypic value with the entire genome. However, in cases where multiple phenotypic traits are correlated, the combined effects of SNPs on these traits may be overlooked using a single-trait GWAS method. In contrast, multi-trait GWAS methods offer higher statistical power and more accurate parameter estimation and location [20]. In this study, the comprehensive GWAS results for both single and multiple traits consistently identified 5 significant SNPs with the genes annotated by these SNPs being FGGY, LHFPL6, OSBPL1A, and NMNAT3. These genes are likely to be potential candidates for pleiotropy related to drip loss in Beijing Black pigs. Among these candidate genes, the FGGY gene encodes a series of carbohydrate kinases, such as fuculokinase, gluconokinase, glycerol kinase, ribulose kinase, and xylulose kinase, which phosphorylates their corresponding sugar substrates [21]. Previous GWAS analyses have identified associations between polymorphisms in this gene and both sporadic amyotrophic lateral sclerosis (ALS) in humans [22] as well as average daily gain (ADG) traits in Nellore cattle [23]. Additionally, recent research has confirmed that FGGY can express at least four alternative splicing variants in myoblasts and play a regulatory role in muscle atrophy through MAPK and PI3K-Akt signaling pathways [24]. Furthermore, studies on long intergenic noncoding genes (lincRNAs) associated with porcine muscle growth have identified FGGY as a potential target gene involved in muscle growth-related processes [25]. After an animal is slaughtered for meat, enzymes in the skeletal muscle metabolize stored carbohydrates into energy through anaerobic glycolysis to maintain energy homeostasis [26]. These biochemical reactions will directly impact the quality of meat, thus highlighting the potential significance of FGGY as a key carbohydrate kinase in postmortem energy metabolism. Moreover, mitochondria can potentially contribute to postmortem metabolism by maintaining their structural and functional integrity for several hours after death, thereby promoting ATP hydrolysis and enhancing glycolytic flux [27]. NMNAT3 is considered as a compensated pathway enzyme for NAD biosynthesis in mitochondria [28]. Lack of NMNAT3 does not affect mitochondrial NAD+ synthesis [29], but the ATP content in skeletal muscle shows an increasing trend under overexpression [30]. This suggests that NMNAT3 may be involved in meat metabolism through mitochondria and ultimately affect drip loss in Beijing Black pigs.
In addition, two candidate genes identified in the GWAS study, namely LHFPL6 and OSBPL1A, are implicated in biological processes related to lipid regulation. LHFPL6 is a member of the lipoma high-speed actin fusion chaperone protein (LHFP)-like family, initially identified as a translocation partner of the high mobility group gene in lipomas [31]. Additionally, a frequent 89 kb deletion in LHFPL6 has been detected in Chinese Simmental cattle and may be linked to average daily gain (ADG) [32]. OSBPL1A belongs to the family of eukaryotic lipid-binding/transfer proteins (LTPs) [33]. It has been demonstrated that OSBPL1A interacts with the small GTPase Rab7 as a sterol-specific regulator to maintain the subcellular distribution of lipids in late endocytic compartments [34]. Moreover, a loss-of-function mutation within OSBPL1A may impair cholesterol removal capacity [35]. Lipids constitute one of the most crucial components in meat. The postmortem oxidation of lipids is a multifaceted process, wherein polyunsaturated fatty acids interact with reactive oxygen species and initiate a cascade of secondary reactions. These secondary reactions also impact the decline in water-holding capacity of meat [36].
Afterward, we conducted a comparative expression profiling analysis on extremely high and low drip loss samples from Beijing Black pigs, resulting in the identification of 880 DEGs. Subsequently, we pinpointed 21 potential DGEs that significantly impact drip loss in Beijing Black pigs, several of which have been previously reported as candidate genes associated with this trait [18]. These DEGs were found to be enriched in pathways associated with extracellular matrix (ECM)–receptor interactions and focal adhesion. Genes involved in these pathways, such as TNC, TNN, ITGA5, and ITGA6, form networks with muscle fibers through binding or adhesion to each other. These processes perform and regulate muscle fiber contraction, which ultimately affects meat drip loss [37]. Besides, there have been numerous reports on the impact of stress-induced heat shock proteins on meat drip loss [38]. The candidate genes in this study contain genes such as HSPB6 and HSPA1L, which may have a pivotal role in regulating cellular osmotic balance under conditions of cellular stress.
To further validate the findings of GWAS, we observed that candidate genes identified by GWAS did not exhibit DGEs in RNA-seq results. This may be attributed to the limited coverage of SNP loci resulting from incomplete genome-wide information provided by the microarray data utilized. Alternatively, genetic variations are often highly correlated with adjacent variants on chromosomes owing to the presence of linkage disequilibrium (LD) phenomenon. Consequently, when analyzing GWAS results, it is common to focus solely on the variation in the region exhibiting the strongest correlation and overlook the true causal variation. Furthermore, this serves to highlight the limitations of inferring causality from GWAS results. Given the pleiotropic effects of genes, the loci identified by GWAS may not necessarily have a direct causal relationship with the associated phenotypes. In this study, the candidate genes FOXO1, OSBPL1A, and DOCK1 identified from GWAS were found to exhibit interactions with the candidate DEGs identified from RNA-seq data through PPI analysis. These findings suggest that these genes may coregulate drip loss traits through mutual regulation. Indeed, the FOXO1 gene is a transcription factor belonging to the forkhead box protein O family, which has been implicated in regulating numerous cellular functions, such as proliferation, metabolism, and differentiation in skeletal muscle [39-41]. Additionally, FOXO1 is believed to play a crucial role in determining the various types of skeletal muscle fibers. Studies have shown that transgenic FOXO1 mice exhibit the altered expression of genes regulating fiber types, which leads to changes in both skeletal muscle mass and function [42]. While DOCK1 functions as a GTPase exchange factor, studies have demonstrated its ability to activate Rac and stimulate actin polymerization in response to signals by a variety of receptors, which is essential for developmental and homeostatic processes, such as myoblast fusion and cell migration [43]. Hence, the aforementioned genes are potential candidates in the interaction between genotype and phenotype affecting drip loss in Beijing Black pigs.
CONCLUSIONS
This study utilizes a combination of GWAS mapping and RNA-seq data to provide evidence of potentially functional SNPs as well as candidate genes associated with drip loss in Beijing Black pigs. Based on the GWAS results, we focus on five SNPs with annotations for four genes (FGGY, LHFPL6, OSBPL1A, and NMNAT3) that were repeatedly detected in single- or multiple-trait GWAS results. These markers are selected as potential pleiotropic indicators associated with drip loss. Furthermore, we have discovered that the genes FOXO1, OSBPL1A, and DOCK1 identified by GWAS exhibit interactions with candidate DGEs obtained from RNA-seq comparative data of Beijing Black pigs exhibiting high and low drip loss groups. Therefore, these genes may influence drip loss traits through gene interaction. Taken collectively, these findings offer crucial insights into the molecular mechanisms underlying drip loss and serve as a theoretical framework for improving meat quality in Beijing Black pigs.
AUTHOR CONTRIBUTIONS
Hongmei Gao: Conceptualization (equal); project administration (equal); writing – original draft (equal). Jingjing Tian: Data curation (equal); funding acquisition (equal). Run Zhang: Data curation (equal); formal analysis (equal). Xiance Liu: Investigation (equal); resources (equal). Hai Liu: Investigation (equal); resources (equal). Fuping Zhao: Software (equal); validation (equal). Zhenhua Xue: Resources (equal). Lixian Wang: Funding acquisition (equal); Supervision (equal); writing – review & editing (equal). Xitao Jing: Methodology (equal); Project administration (equal); resources (equal); visualization (equal); writing – review & editing (equal). Longchao Zhang: Conceptualization (equal); supervision (equal).
ACKNOWLEDGEMENTS
The authors thank all the members of our laboratory for their practical, cordial, and selfless support in writing this thesis. This research was supported by the National Key R&D Program of China (2021YFD1301101), National Swine Industry Technology System (CARS-35), and Agricultural Science and Technology Innovation Program (ASTIP-IAS02).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS STATEMENT
All animals were treated humanely according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” published by the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China). The experimental protocols used in this study were approved by the Animal Care and Use Committee (No. IASCAAS-PG-39).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article and its additional information files.




