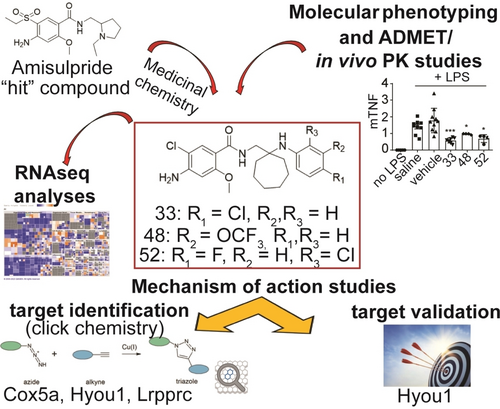
Discovery of the First-in-Class Inhibitors of Hypoxia Up-Regulated Protein 1 (HYOU1) Suppressing Pathogenic Fibroblast Activation
Graphical Abstract
A multidisciplinary pipeline consisting of medicinal chemistry, molecular phenotyping, chemoproteomics, RNA-sequencing, short hairpin RNAs (shRNAs)-mediated gene silencing and in vivo studies led to a novel series of compounds suppressing pathogenic fibroblast activation via Hypoxia up-regulated protein 1 (HYOU1) inhibition. The first reported HYOU1 inhibitors are presented as a promising therapeutic approach in fibroblast-related diseases.
Abstract
Fibroblasts are key regulators of inflammation, fibrosis, and cancer. Targeting their activation in these complex diseases has emerged as a novel strategy to restore tissue homeostasis. Here, we present a multidisciplinary lead discovery approach to identify and optimize small molecule inhibitors of pathogenic fibroblast activation. The study encompasses medicinal chemistry, molecular phenotyping assays, chemoproteomics, bulk RNA-sequencing analysis, target validation experiments, and chemical absorption, distribution, metabolism, excretion and toxicity (ADMET)/pharmacokinetic (PK)/in vivo evaluation. The parallel synthesis employed for the production of the new benzamide derivatives enabled us to a) pinpoint key structural elements of the scaffold that provide potent fibroblast-deactivating effects in cells, b) discriminate atoms or groups that favor or disfavor a desirable ADMET profile, and c) identify metabolic “hot spots”. Furthermore, we report the discovery of the first-in-class inhibitor leads for hypoxia up-regulated protein 1 (HYOU1), a member of the heat shock protein 70 (HSP70) family often associated with cellular stress responses, particularly under hypoxic conditions. Targeting HYOU1 may therefore represent a potentially novel strategy to modulate fibroblast activation and treat chronic inflammatory and fibrotic disorders.
Introduction
Fibroblasts are stromal cells that play pivotal roles in wound healing, tissue repair, and extracellular matrix (ECM) production, while also being involved in various disease processes such as immunity, inflammation and cancer.1 In terms of immunity and inflammation, fibroblasts have been shown to play a key role in the innate immune response by producing cytokines and chemokines that recruit and activate immune cells. Especially in chronic inflammatory diseases (CIDs) that often lead to fibrosis, fibroblasts can become overactive and produce excessive amounts of ECM components, leading to tissue damage, scarring, and progressively to organ failure.
Current treatments for CIDs like rheumatoid arthritis (RA), inflammatory bowel disease and psoriasis primarily focus on regulating the immune response and the elevated cytokines or activated molecular pathways (e.g. anti-TNF/IL-6 biologics).2 Furthermore, the anti-fibrotic drugs available today (pirfenidone, and nintedanib) mainly alleviate fibrosis’ symptoms without directly addressing fibroblast pathogenicity. Targeting fibroblasts could therefore offer an alternative therapeutic approach for CIDs and fibrosis, particularly for patients experiencing side effects or resistance to conventional immunosuppressive treatments.3, 4, 5 Accordingly, this study aims at a) discovering novel small molecules that inhibit pathogenic fibroblast activation, and b) exploring new therapeutic mechanisms.
Our previous research demonstrated that the antipsychotic drug amisulpride exhibits significant activity against chronic polyarthritis in the hTNFtg mouse model (overexpressing human tumor necrosis factor, TNF) by suppressing the activation of joint synovial fibroblasts, the key pathogenic drivers in this RA animal model.6 Using amisulpride as a “hit,” we have now developed a new series of bioactive compounds targeting various pathogenic fibroblast activation properties, such as their inflammatory secretome, wound healing potential and ECM-related gene expression (Scheme 1). The latter was accompanied by a potent anti-inflammatory effect of the compounds in vivo, effectively mitigating acute lipopolysaccharide (LPS)-mediated endotoxemia in mice. Importantly, hypoxia up-regulated protein 1 (HYOU1) was identified as the primary molecular target of the new derivatives, thus rendering them the first-in-class HYOU1 inhibitors. Overall, these results highlight the potential of these compounds as a novel promising therapeutic strategy with broad applications in the treatment of various diseases involving pathogenic fibroblast activation (e.g. CIDs, fibrosis, and cancer).
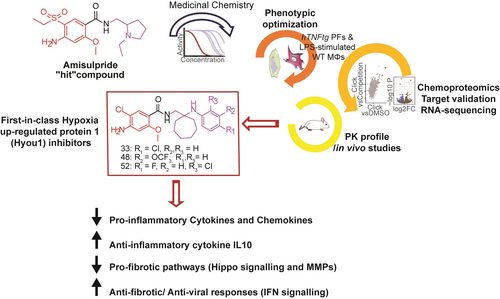
Overview of the present study highlighting the discovery of the first-in-class HYOU1 inhibitors against pathogenic fibroblast activation.
Results and Discussion
Discovery of Novel Small Molecules Suppressing the Inflammatory Potential of Activated Fibroblasts
To discover new “leads”, we initially developed a phenotypic screening assay in hTNFtg primary joint fibroblasts (PFs), used here as a model of activated fibroblasts. This cellular context exhibits an inflammatory and destructive phenotype, greatly mimicking human RA fibroblasts.7, 8 The compound's inhibitory effect against the pro-inflammatory and pathogenic chemokine levels CCL20 (MIP3) and CCL5 (RANTES) was used as a primary readout. Both chemokines are strongly induced by TNF,9 a cytokine regulating fibroblast pathogenicity in multiple diseases.10 Additionally, we used wild-type (WT) PFs exogenously stimulated by TNF, aiming at accessing in parallel the effect of each compound on both acute (hTNF-induced WT-PFs) and chronic (hTNFtg-PFs) TNF-driven cell responses.
First, we developed a synthetic route that allowed the parallel synthesis of derivatives for the exploration of molecular phenotyping studies (Figure 1A). Using amisulpride as a “hit”, we replaced its N-ethylpyrrolidinemethyl- and 4-amino-5-(ethylsulfonyl)-2-methoxy-groups with different substituted amines and (hetero)aromatic carboxylic acids (Table ST1), respectively. Compound 18 emerged from this preliminary exploration, showing a 6–50-fold better effect than amisulpride in reducing MIP3 and RANTES levels (Figure S1, Table ST1).
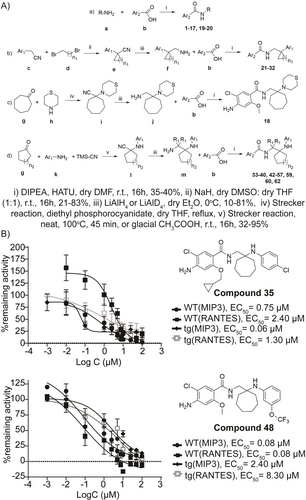
A) General routes followed for the synthesis of the compounds of the new series. B) Effect of 35 and 48 on MIP3 and RANTES levels in hTNF (1 ng/mL)-induced WT-synovial PFs and in hTNFtg PFs. All data are shown as a mean±SEM and EC50 is calculated as a mean of three biological replicates.
Replacement of the thiomorpholine ring of 18 with (hetero)aromatic moieties (22–26, Figure S2, Table ST2) favored an increase in cellular potency, with 21, 22 and 24 being the most active compounds. The pyridine isostere of 21 (27) exhibited a 30-fold better reduction in hTNFtg-MIP3 levels, while the activity was abrogated by the bicyclic chromane derivative 28 (Figure S1, Table ST2).
Next, we assessed the impact of the cycloalkyl ring size on potency by synthesizing and testing compounds 29–32 (Figure S2, Table ST2). The cyclopentyl ring (31) seems to preserve cellular activity, while the cyclobutyl (30) and cyclohexyl (32) groups decrease the overall effect and the cyclopropyl group (29) is not tolerated.
Compound 21 was then subjected to a preliminary characterization of main physicochemical properties potentially affecting the “drug-like” profile of a bioactive molecule. It exhibited high solubility in simulated intestinal fluid (SIF) and no detected cytotoxicity, cardiotoxicity and genotoxicity (Table ST3). However, this was counterbalanced by its very low aqueous solubility in simulated gastric fluid (SGF) and in phosphate buffer (PBS, pH 7.4), as well as by its fast metabolic clearance.
To expand the chemical space of this series further, we increased the flexibility of the scaffold by introducing an amino-linker between the cycloheptane and the aromatic ring. This structural modification (33) conferred a better ADMET profile compared to the respective rigid analogue 21 (Table ST3), while preserving cellular potency (Figure S2, Table ST4).
The impact of the methyl group of the methoxy unit of 33 on activity was then investigated. We found that although the ethyl group (34) is not tolerated, substitution with cyclopropylmethyl (35, Figure 1B) or isopentyl (36) group offers a better anti-inflammatory effect compared to that of 33 (Figure S2, Table ST4). Moreover, this substitution significantly reduced the metabolic clearance ex vivo but at the expense of higher lipophilicity, lower permeability, lower aqueous solubility in SGF and in PBS (pH 7.4) and higher plasma protein binding (35, Table ST3). What is more, the chlorine substitution of 4-amino-5-chloro-2-methoxyphenyl group seems to be crucial for cellular activity since its replacement with other atoms (−Br, 37) or aromatic groups (4-fluorophenyl, 38) abrogated potency (Figure S2, Table ST4).
The last round of structural modifications performed was at the aromatic substitution of the amino-linker of 33 (Figure S3, Table ST5). Substituents endowed with molecular diversity were incorporated to assess the impact of size, shape, rigidity and electrostatic complementarities on activity. It can be inferred that lipophilicity at this position is of essence as polar groups (41–44, 46, 55–58, Figure S3, Table ST5) significantly reduced potency. 40, 48 and 52 which emerged from this exploration as the most potent analogues (Figures 1B, S3, Table ST5), were evaluated for their ADMET profile. Compared to 33, both 48 and 52 showed similar lipophilicity, higher plasma protein binding, lower aqueous solubility, especially in SGF and PBS (pH 7.4), and higher metabolic clearance rate. Moreover, although the aromatic substituent of 40 provides better cell membrane permeability and lower plasma protein binding, these are compromised by lower solubility in PBS (pH 7.4), a higher metabolic clearance rate and an increased human ether-a-go-go related gene (hERG) inhibition, a prelude of cardiotoxicity (Table ST3).
From the aforementioned studies, 33 was prioritized, among the analogues tested, for further pharmacological, in vivo and mechanistic studies since it significantly decreased MIP3 and RANTES levels in both acute (hTNF-induced WT-PFs) and chronic (hTNFtg-PFs) activation of PFs (Table ST4) without being cytotoxic, even at high concentrations (Figure S4, Table ST3). Furthermore, it showed high solubility in SGF and SIF, moderate permeability as well as solubility in PBS (pH 7.4) and desirable lipophilicity (Table ST3), while its fast metabolic clearance is subject to further optimization. For consistency, two of the most potent derivatives in cells (48, 52) were also included. Despite the low solubility and permeability (Table ST3) displayed by 48 and 52, their potent cellular effect (Table ST5) could be attributed to a high binding affinity against the target protein, compensating for their inferior physicochemical profile.
Identification of Metabolic “Hot Spots”
In addition, since the main problem of the majority of this class of compounds was the low metabolic stability leading to high clearance, we attempted to pinpoint the metabolic “hot spots” of 33’s structure by synthesizing and testing 59–62 (Table ST6). Compound 35, showing the best metabolic stability among the analogues tested (Table ST3) was also included for comparison. The metabolically labile methoxy group of 33 (prone to demethylation) was replaced by cyclopropylmethoxy in 35 and oxetan-2-yl-methoxy in 59. The cycloheptane ring (prone to oxidation) was replaced by the more metabolically resistant cyclopentane in 60 and spiro[2.3]hexane unit in 61, while in 62 the hydrogens of the methylene group which are prone to oxidation were substituted by the heavy isotope deuterium. Apart from the cyclopropyl group (35), all the other modifications either decreased (60, 62) or abolished potency (59, 61), especially in hTNFtg-PFs (Figure S5, Table ST6). However, the metabolic clearance rate and the elimination half-time were significantly improved only in 35 and 60 (Table ST3), clearly showing that primarily the demethylation of the methoxy group and secondarily the oxidation of cycloheptane ring are the metabolic “hot spots” of 33. The fast metabolic clearance observed in 59 could be attributed to the hydrolytic ring opening of the oxetane ring by human microsomal epoxide hydrolase.11 The structural liabilities determined in this study are driving our current efforts to develop optimized molecules of this series with a balanced potency and “drug-like” profile. These efforts will be published elsewhere.
Anti-Inflammatory Potential
Compounds 33, 48 and 52 were further evaluated for their ability to reduce pivotal pro-inflammatory chemokines/NF-kB-targeted genes in hTNFtg-PFs and LPS-treated WT-macrophages (MΦs). Both cellular settings produce various cytokines/chemokines that contribute to inflammatory and pro-fibrotic responses.
The compounds effectively reduced the production of chemokines involved in T-cell (e.g. macrophage inflammatory protein-3 MIP3) and monocyte recruitment (e.g. monocyte chemoattractant protein-1 MCP-1, MIP-1β, RANTES), as well as those promoting neutrophil recruitment (e.g. keratinocyte-derived chemokine KC, LPS-induced CXC chemokine LIX) and cell migration (e.g. EOTAXIN) of activated hTNFtg-PFs. Similar effects were observed in LPS-treated WT-MΦs, indicating a broad anti-inflammatory effect of the new analogues (Figure 2A, S6). Additionally, the compounds decreased significantly the levels of the cytokine Interleukin 6 (IL-6, that is abundant in CIDs and fibrosis and associated with disease severity)12 in a dose-dependent and non-toxic manner (Figure S4), with 52 exhibiting the strongest effect (Figure 2B).
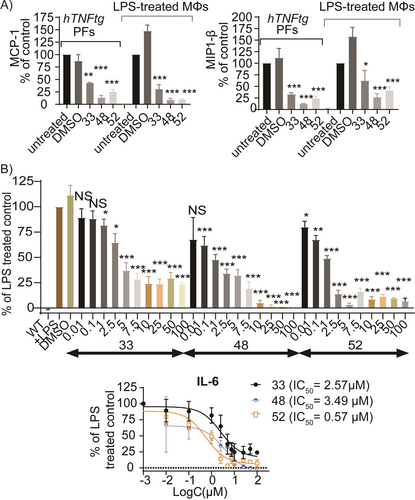
A) Pro-inflammatory chemokine levels (MCP-1 and MIP-1β) upon treatment of hTNFtg-PFs and LPS (10 ng/mL)-induced WT-MΦs with 33, 48 and 52 (10 μΜ). B) Dose-response of 33, 48 and 52 against IL-6 levels in LPS (10 ng/mL)-treated WT-MΦs. All data are shown as a mean±SEM of six biological replicates and all comparisons are made against DMSO control using one-way ANOVA (Dunnett's multiple correction method), *** p<0.001, ** p<0.01, * p<0.05, NS not significant.
Mechanism of Action in Cells and In vitro
To explore further the impact of the new series on pathogenic fibroblasts, we evaluated 33, 48 and 52 for their ability to inhibit the invasive phenotype of activated PFs. Treatment with these compounds did not affect fibroblast proliferation, as evidenced by unaffected cell cycle progression in hTNFtg- and WT-PFs (Figure S7). In contrast, all derivatives demonstrated a significant inhibition of hTNFtg-PFs motility in the wound healing assay. Notably, the presence of the cycloheptane ring and the amino-linker are determinants for anti-migratory activity, since the 6-membered ring and rigid analogue of 33 (32) showed no effect (Figure S8). The concentrations used in these experiments are non-toxic as demonstrated by Annexin staining (Figure S9).
Furthermore, representative active analogues (24, 33, 48, 52) were tested for their ability to inhibit TNF-mediated cytotoxicity in L929 cells (Figure S10). 24 and 48 exhibited significant inhibition (IC50 values of 49.2 and 23.5 μM, respectively), comparable to the TNF inhibitor C87 which is available on the market (IC50=10 μM),13 used as a reference control. Overall, the new series primarily affects the migration rather than the proliferation of activated PFs, while some derivatives can effectively block TNF-mediated signaling.
Our focus then shifted to understanding the molecular mechanism underlying the suppressive effect of the compounds on activated fibroblast responses. We assumed that the mode of action of the new analogues differentiates from that of amisulpride,6 based on two observations: a) the inhibitory effect of amisulpride on different chemokines was found to be much weaker compared to the new compounds (Table ST1),6 and b) unlike the new series, amisulpride did not inhibit the migration of activated PFs even at high concentrations (Figure S8). Accordingly, we tested 33 and 52 for their dopaminergic activity in vitro, particularly against D2R/D3R dopamine receptors, the main targets of amisulpride. Neither compound acted as an agonist nor antagonist of D2R/D3R receptors (Figure S11), indicating that the new derivatives lack the antipsychotic effects associated with the parent drug.
We then utilized SPD304 as a TNF reference antagonist, to investigate whether 24, 33, 48 and 52 could disrupt binding of TNF to its main receptor TNF receptor I (TNFRI) by an in vitro ELISA assay.6, 14 Unlike SPD304,15 which fully blocked TNF/TNFRI interaction (IC50=8.7 μM) as expected, all new derivatives acted as partial TNF antagonists (Figure S12). However, due to their weak effect, the inhibition of TNF-TNFRI binding cannot be considered as the primary molecular mechanism of the new compounds.
Target Identification and Prioritization
To dissect the molecular mechanism of action of the new series, we then focused on deploying chemoproteomic approaches using hTNFtg-PFs as a pathogenic cell target. To this end, we designed and synthesized two click probes for each of compounds 33 (Clicks 1, 2, Figure 3A) and 52 (Clicks 3, 4, Figure 3A). The click tag was incorporated either in the methoxy group (Clicks 1, 3) or at the 5-position of the aromatic ring (Clicks 2, 4) of both molecules.
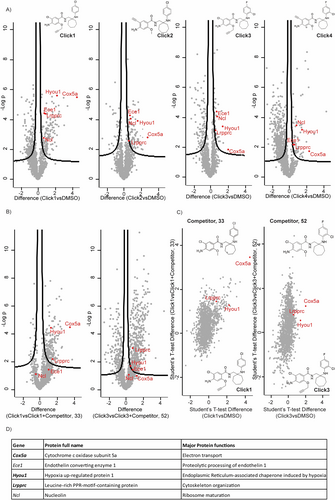
Α) Volcano plots of proteins enriched by Click 1–4 probes (25 μΜ) versus no probe (DMSO) in hTNFtg-PFs. Common enriched proteins between all clicks vs DMSO control are highlighted. B) Volcano plots of proteins enriched by probes Click 1 and Click 3 (25 μΜ) versus the respective competition control [Click 1 (25 μΜ) and Competitor 33 (500 μΜ), and Click 3 (25 μM) and Competitor 52 (500 μΜ)] in hTNFtg-PFs. C) Protein enrichment by probe Click 1 compared to no probe (DMSO) and competition control (Click 1 and Competitor 33), and protein enrichment by probe Click 3 compared to no probe (DMSO) and competition control (Click 3 and Competitor 52) in hTNFtg-PFs. D) Common top hits derived from A), and in bold from B) and C). Each sample was analyzed in three technical replicates.
After confirming that the inhibitory activity of Clicks 1–4 against MIP3 and RANTES levels (Table ST5) occurred over a dose-response range similar to that of the parent derivatives 33 and 52 (Tables ST4, ST5), hTNFtg-PFs were treated with each click derivative (Clicks 1–4, 25 μM). Three controls were used alongside. The first control involved vehicle (DMSO) treated cells, while the other two (competition) involved treatment of hTNFtg-PFs with 25 μM of Click-1 or Click-3 along with a 20-fold excess concentration of their precursors 33 or 52, respectively, to block the enrichment of active probe binding proteins. The use of four chemically diverse click probes and three controls in the experiment aimed at excluding as many artifacts (non-specific hits) as possible during LC–MS/MS analysis, enabling us to identify a limited number of potential primary protein targets for validation.
Following cell lysis, the lysates were subjected to the Cu+-catalyzed alkyne-azide cycloaddition for conjugating target proteins with a biotin tag. The biotin-tagged proteins were then pulled down on streptavidin beads. After tryptic digestion, the peptides were analyzed by LC–MS/MS.
Volcano plots of enriched proteins by Click 1–4 probes versus the DMSO group highlighted five common candidates that were highly significant in all 4 groups (Figures 3A, 3D). Then, to distinguish therapeutically relevant targets from non-specific or irrelevant hits we initially compared all proteins that were highly enriched by Clicks 1–4 vs DMSO with those enriched by active Click 1 and Click 3 probes but displaced by the competitors 33 (Click 1+33 group) and 52 (Click 3+52 group), respectively (Figure 3B). This comparison showed high statistical significance in both cases (three technical replicates, FDR=0.05) that 33 and 52 are able to suppress the pull down of three proteins (Figure 3B, Figure 3D in bold), with the competition effect being much stronger in 33 compared to 52 (Figure 3B). The suboptimal competition provided by 52 could be attributed to its low aqueous solubility and permeability (Table ST3), which both decrease the compound's available concentration to effectively compete with the Click 3 probe inside the cells. Lastly, combining differences in proteins identified in active Click 1–4 probe samples but not in the three controls (DMSO/Click 1+33/Click 3+52, Figure 3C) corroborated the shortlist of the 3 potential targets (Figure 3D in bold).
To determine the protein(s) to prioritize in target validation experiments, a bulk-RNA sequencing analysis was performed to identify pathways differentially regulated post compound exposure (Figure 4). The expression profiles of diseased hTNFtg-PFs treated with 33 for 48 hours were compared to untreated cells (DMSO control). 33 inhibited multiple pathways found activated in pathogenic fibroblasts, including extracellular matrisome alteration, TNF signaling, cytokine/chemokine production and collagen fibril organization (Figures 4A, 4C). Pathway enrichment analysis revealed that 33 also attenuated Hippo signaling and Hypoxia inducing factor 1 (HIF-1)-mediated hypoxia resistance (Figures 4A, 4C). The HIF-1 pathway regulates fibroblast adaptation in hypoxic conditions of hyper-proliferating environments of chronic diseases,16 while Hippo signaling is involved in fibrotic and cancer responses.17 Additionally, 33 activated interferon (IFN) signaling (Figures 4A, 4C), indicating the compound's potential to support antiviral and antifibrotic responses.18 Interestingly, IFNγ has been shown to enhance the effects of the anti-fibrotic drug pirfenidone by reducing lung fibroblast activation and differentiation.19 Finally, the expression of Mmp3 and Bmp2 genes, both being highly upregulated in activated fibroblasts promoting chronic inflammation and fibrosis, was confirmed to be downregulated by 33 (as well as by 48 and 52) when compared to the untreated control (Figure 4B). Ingenuity pathway analysis (IPA)20 indicated further the potential involvement of the derivatives in pathways and conditions related mostly to fibrosis and cancer (Figure S13), thus suggesting their relevance in fibroblast-mediated disease contexts.
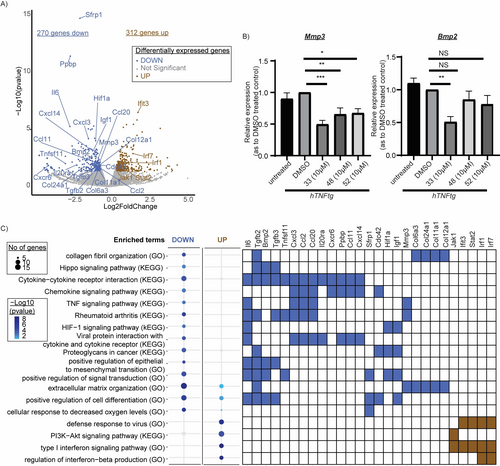
A) Volcano plot indicating deregulated genes upon 33 treatment (10 μΜ, 48 h) versus untreated control (DMSO). Genes of interest are highlighted. B) Validation of representative fibroblast activation genes expression (Mmp3, Bmp2) upon compound 33 treatment (10 μΜ, 48 h). C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) biological processes enriched in the upregulated and downregulated genes upon treatment with 33 (10 μΜ, 48 h). Α Gene versus Function heatmap indicates genes of interest belonging to the selected functional categories. All data are shown as a mean±SEM of three biological replicates and all comparisons are made against DMSO control using one-way ANOVA (Dunnett's multiple correction method), *** p<0.001, ** p<0.01, * p<0.0.
Comparing the RNA-sequencing (Figure 4C) and IPA analysis (Figure S13) with literature data about the three potential protein targets (Figure 3D in bold), we concluded that 33’s gene expression signature most closely matched that of HYOU1 inhibition, since HYOU1 is known a) to be involved in chronic inflammation, stimulating production of various pro-inflammatory mediators,21-23 b) to be highly up-regulated, together with HIF-1α, in hypoxic conditions,24, 25 c) to play a key role in cancer22-24, 26, 27-29 and fibrosis,30, 31 while its inhibition d) reinforces the defense response to virus,32 and e) results in Interferon signaling pathway upregulation.28 Accordingly, HYOU1 was advanced to target validation studies.
HYOU1 (GRP170/ORP150) is a member of the non-canonical heat shock proteins “HSP70 Super-Family”, residing in the endoplasmic reticulum and functioning as a molecular chaperone. It aids in protein folding and transportation of secretory or transmembrane proteins. It has been shown to exert immunoregulatory activities in certain immunopathologies,22, 23 while its expression is found increased under stress conditions (e.g. reductive stress, anoxia).22 However, its specific role on pathogenic fibroblast activation remains unexplored.
Target Validation
To validate the function of HYOU1 in the pathogenic activation of fibroblasts, short hairpin RNAs (shRNAs) were designed, and the Lenti-X Lentiviral expression system was employed as a delivery system to knock down Hyou1 in hTNFtg-PFs. In addition, Cox5α silencing was included for comparison purposes.
Successful downregulation of each mRNA compared to the scramble-treated control was confirmed (Figures 5Α, Β) and the impact of gene silencing on cell viability was assessed (Figure S14a). Hyou1 downregulation resulted in a significant reduction of both MIP3 (Figure 5E) and RANTES levels (Figure 5F), while Cox5α downregulation had no effect on the respective chemokine levels (Figures 5C, D). Hyou1 deletion also led to a decreased expression of other pro-inflammatory chemokines (Figures 5G–K), resulting in an anti-inflammatory effect similar to that of compounds 33, 48 and 52 (Figures 1B, 2A, S2, S3, S6). Furthermore, since HYOU1 serves as a molecular chaperone for matrix metalloproteinase-2 (MMP2) secretion, promoting tumor invasion in malignancies,33 we investigated whether a correlation between Hyou1 and Mmp2 expression could exist in PFs. Figure 5L shows that Hyou1 downregulation significantly reduced Mmp2 expression, which is also known to be involved in fibroblast-to-myofibroblast transition. Similarly, treatment of hTNFtg-PFs with 33, 48 and 52 notably inhibited Mmp2 levels (Figure 5M).
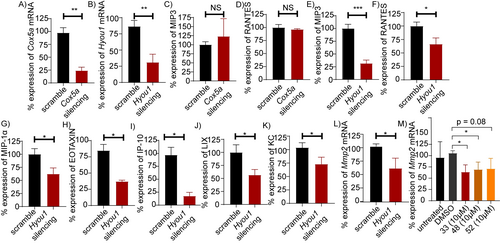
ShRNAs-mediated silencing of Cox5α and Hyou1 genes in hTNFtg-PFs using the Lenti-X Lentiviral expression system: A) % Cox5α and, B) % Hyou1 expression levels. C) Effect of Cox5α downregulation on MIP3 and D) RANTES expression levels. Effect of Hyou1 downregulation on E) MIP3, F) RANTES, G) MIP-1α, H) EOTAXIN, I) IP-10, J) LIX, K) KC, and L) Mmp2 expression levels. M) Mmp2 expression upon 33, 48 and 52 (10 μΜ) treatment. All data are shown as a mean±SEM of nine biological replicates and all comparisons are made against hTNFtg-scramble sample (Figures 4A–L) or hTNFtg-DMSO (Figure 4M), using Student's t-test, *** p<0.001, ** p<0.01, * p<0.05, NS not significant.
Lastly, Gossypolacetic acid (GAA), a specific LRPPRC inhibitor effectively inducing LRPPRC degradation,34 was used as a pharmacological tool to evaluate the anti-inflammatory effect of LRPPRC inhibition in PFs. GAA was found to be inactive against MIP3 and RANTES in hTNFtg-PFs (Figure S15), thus implying no involvement of LRPPRC in pathogenic fibroblast activation.
Altogether, these target validation results indicate that the compounds of the new series exert dominant inhibitory effects on PF activation through HYOU1 targeting.
Since no in vitro assay for evaluating the binding affinity of ligands to HYOU1 has been reported in the literature, we then characterized the physical interaction between 33 and HYOU1. We performed a cellular thermal shift assay (CETSA)35a by treating hTNFtg-PF lysates with 33, heating as indicated (Figure 6A), and probing the remaining soluble protein by western blotting using a specific anti-HYOU1 antibody. We observed a thermal stabilization of HYOU1 in 33-treated PFs when compared to the vehicle (DMSO)-treated PFs, indicating a direct interaction (Figures 6A, 6B). In parallel, and considering that the full length HYOU1 protein has not been expressed and purified so far, the ability of 33 to bind to the commercially available, truncated HYOU1 recombinant protein (protein sequence at positions 36-583) was evaluated by Differential Scanning Fluorimetry (DSF).35b A substantial difference in thermal stability of 4 °C was noticed for the HYOU136-583-33 complex, attributed to the protection of the protein from thermal denaturation in the presence of 33 (Figure 6C).
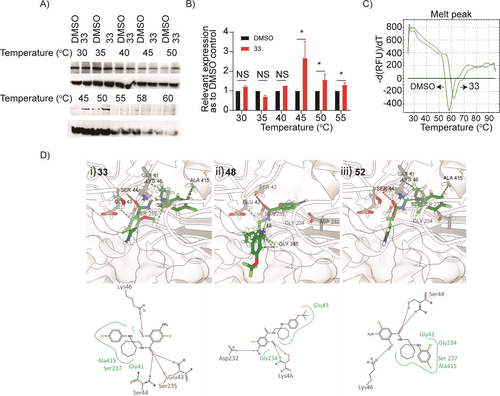
A) Representative western blots showing thermostable HYOU1 following indicated heat shocks (30–60 °C) in the presence of DMSO or compound 33 at 25 μΜ in target engagement experiments in hTNFtg PFs using CETSA (uncropped gel images can be found in supplementary Figure S14b). B) Quantification of target engagement experiments described in A), by plotting the ratio of HYOU1 expression compared to ACTIN loading control, setting in each temperature the relative DMSO control as 1. All data are shown as a mean±SEM of three-six biological replicates and all comparisons are made against each relevant DMSO sample, using Student's t-test, ***p<0.001, **p<0.01, *p<0.05, NS not significant. C) Differential Scanning Fluorimetry (DSF)17 denaturation curves of HYOU136-583 (10 μΜ) in the presence of DMSO or compound 33 (20 x), d (RFU)/dT: differential (Relative Fluorescence Units)/ differential Temperature. D) Calculated binding modes of i) 33, ii) 48 and iii) 52 in HYOU1’s homology model produced with SWISS-MODEL37 and Modeller 10.2.38, 39 Figure was prepared using ChimeraX40-42 and PoseView.43, 44
Finally, a comprehensive analysis was conducted, combining cheminformatics and molecular docking calculations, which involved generating a homology model of the mouse HYOU1 protein. The calculations gave a supportive result, complementary to the previous pharmacological experiments, providing insights into potential binding sites for the most studied and validated compounds 33, 48 and 52 within HYOU1. A detailed account of the HYOU1 modeling, ligand docking, and binding energy calculations will be published elsewhere. The docking calculations revealed that all compounds exhibited favorable binding affinities to the selected HYOU1 binding site (Table ST7). Examination of the docked compounds conformations reveals that they are positioned within hydrogen bond length of essential residues in the HYOU1 mouse protein. These include Glu43, Ser44, Lys46, Asp232, Ser235, and Ser237, which is consistent with the interaction pattern observed for the adenosine segment of the native ligand adenosine diphosphate (ADP).36 33 is in hydrogen bonding distance with Ser44, Ser235 and Glu43, with its chlorobenzene and cycloalkyl moieties being in close contact with the hydrophobic residues Ala415, Gly41 and Ser237 (Figure 6Di). 48 potentially forms hydrogen bonds between its 4-amino-5-chloro-2-methoxyphenyl group and Asp232, while it is within the proximity of Glu43 and Gly234 (Figure 6Dii). Lastly, 52 is stabilized by Ser44 and Lys46, with its chloro-fluoro benzene moiety facing inward, as the chlorobenzene group of 33, towards Gly41, Gly234, Ser237 and Ala415 within the HYOU1 pocket (Figure 6Diii).
In Vivo Activity of the New Series
Prior to assessing the in vivo effect, and considering its high ex vivo metabolic clearance rate (Table ST3), we performed a Pharmacokinetic study (PK) for 33 to determine an appropriate per os dose that would ensure sufficient compound exposure to the systemic circulation. This evaluation would also allow us to assess the therapeutic potential of HYOU1. Intravenous (iv) administration of a single dose of 4.5 mg/kg in C57/BL6 mice revealed a favorable volume of distribution (Vd), associated with a rapid half-life distribution time. However, consistent with the ex vivo ADMET studies (Table ST3), an increased rate of in vivo metabolic clearance was observed (Figure 7, S16), accompanied by a relatively short elimination half-life time (t1/2). Considering both iv PK and ADMET data (Figure 7, Table ST3), we assessed the oral exposure of 33 at an increased single dose of 100 mg/kg (Figure 7). At this dose, 33 demonstrated a rapid entry into the circulation, reaching maximum blood concentration (tmax) at 1 hour while extending its t1/2 to 1.83 hours. The oral bioavailability (F%) of 33 at this dose was 62.1 % (Figure 7).
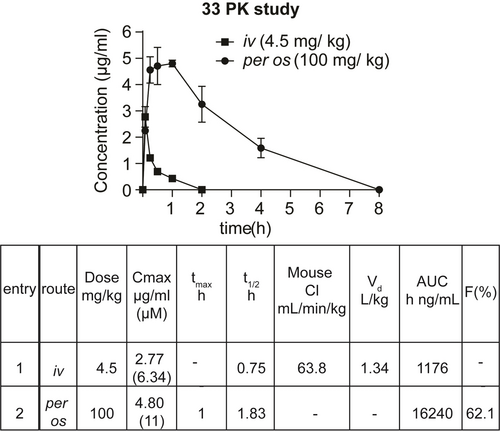
Pharmacokinetic (PK) profile of compound 33 in male C57/BL6 mice after iv (4.5 mg/kg) and per os (100 mg/kg) administration. Saline : captisol (85 : 15 ratio) was used as a vehicle. Data are presented as a mean±SEM of two biological replicates/time point (tmax: maximum blood concentration time, t1/2: half-life time, Cl: Clearance, Vd: distribution volume, AUC: area under curve, F: oral bioavailability).
Then, based on the PK profile of 33 (Figure 7), the in vivo anti-inflammatory effect of 33, 48 and 52 was evaluated. The compounds were orally administered at a dose of 100 mg/kg together with the intraperitoneal (ip) administration of LPS to induce the inflammatory response. Serum levels of the pro-inflammatory cytokines TNF and IL-6 were measured at 1.5 (close to the tmax of 33) and 6 hours (when low compound levels are detectable in plasma, Figure 7) post-dose. All tested derivatives decreased TNF levels at both time points (Figure 8), whereas IL-6 levels were reduced at 6 hours post-administration (Figure S17). The potential of this novel series to exhibit a multifunctional anti-inflammatory profile was also assessed by testing the in vivo activity of 33 on a panel of different inflammatory mediators. Interestingly, 33 significantly decreased the levels of various cytokines (e.g. IL-1β, IL-27, IL-12p70, granulocyte macrophage colony-stimulating factor GM-CSF, interferon gamma-induced protein-10 IP-10 and KC) and chemokines (e.g. RANTES, MIP-1α, MIP-1β, MCP-1 and macrophage-derived chemokine MDC) after LPS stimulation (Figure 8, S18). In contrast, the levels of the anti-inflammatory cytokine IL-10 were increased (Figure 8).
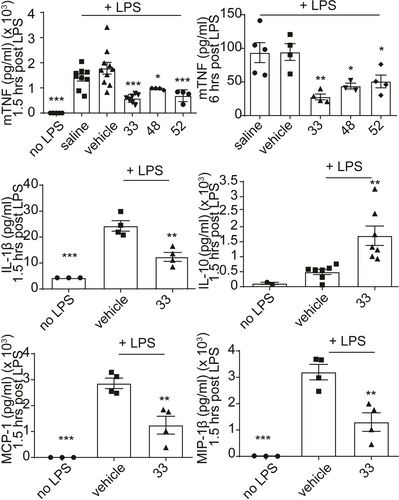
In vivo effect of 33, 48 and 52 on reducing mTNF levels (1.5 and 6 h post-dose) and effect of 33 on other pro-inflammatory mediators (1.5 h post-dose) in the LPS-induced endotoxemia mouse model. All compounds were administered per os (100 mg/kg) together with the ip administration of LPS (1 μg/mouse) in C57/BL6 mice. Saline : captisol (85 : 15 ratio) was used as a vehicle. All data are shown as a mean±SEM of four-eight biological replicates and all comparisons were made against vehicle-treated control using one-way ANOVA (Dunnett's multiple correction method), ***p<0.001, **p<0.01, *p<0.05, NS not significant.
Conclusion
Fibroblasts have recently emerged as highly important pathogenic mediators of immunity, inflammation, and cancer.1, 45 In this study we aimed at translating this knowledge into new effective therapeutics by identifying new compounds capable of deactivating pathogenic fibroblasts. We have previously shown that the atypical antipsychotic drug amisulpride is able to modify the activated phenotype of joint synovial fibroblasts and improve the clinical score of polyarthritis.6 Setting amisulpride as a starting point, we conducted phenotypic optimization studies to develop novel molecules exhibiting anti-inflammatory and anti-fibrotic properties. To this end, we used pathogenic primary fibroblasts activated by TNF in both acute and chronic settings. The most potent derivatives were further evaluated for their ADMET properties to refine molecules exhibiting an acceptable activity/“drug-like” profile, whereas metabolic “hot spot” identification studies were also performed. Compound 33 emerged from these preliminary efforts, displaying a potent effect across a panel of pro-inflammatory cytokines and chemokines, a high solubility in SGF and SIF, a moderate permeability and solubility in PBS (7.4) and no toxicity issues. However, it also exhibited a fast intrinsic clearance, the improvement of which is the goal of our current efforts. To gain insights into the molecular mechanism of this new series of molecules, representative derivatives underwent chemoproteomics studies followed by RNA-seq and target validation analysis. Lastly, 33 was used as a pharmacological tool to further assess in vivo the therapeutic potential of the new target identified.
Our study establishes for the first time the potential of HYOU1 as a therapeutic target in CIDs. Various lines of evidence implicate HYOU1 in several biological processes relevant to disease. For example, proteomic analysis of synovial samples from patients with osteoarthritis, chronic pyrophosphate arthropathy, and RA demonstrates a significant correlation between HYOU1 expression and increased histological inflammatory scores.21 Additionally, extracellular HYOU1 has been identified as an alarmin in response to tissue damage, amplifying inflammatory responses triggered by microbial signals and damage-associated molecular patterns (DAMPs).22, 23 Furthermore, HYOU1 is linked to viral IL-6 (vIL-6), promoting pro-inflammatory signaling, survival, and migration of endothelial cells.32
Importantly, HYOU1 is upregulated in hypoxic conditions and cancer, playing a critical role in various malignancies by supporting tumor cell survival, while its overexpression promotes cytokine (such as TNF) production from tumor immune cells. It also favors epithelial to mesenchymal transition (EMT) by acting as a chaperone to both vascular endothelial growth factor (VEGF) and MMP2, thus facilitating the invasive phenotype of tumor cells.24 As a result, the potential of HYOU1 ligands as promising anticancer agents has been extensively mentioned over the last decade.22-24, 26, 27-29
Similarly, HYOU1 has been shown to stimulate idiopathic pulmonary fibrosis (IPF) in mouse models by increasing TGF-β1 levels and myofibroblasts in the lungs.30 It is worth noting that hyou1 is among the top differentially expressed genes in multi-organ fibrosis analyses, including the heart, lung, liver, kidney, and pancreas.31
Although further research is needed to fully comprehend the precise role of HYOU1 in chronic diseases, its activity appears to be primarily driven by its chaperoning property that controls protein quality and degradation.29 This is evidenced by its ability to regulate MMP2 secretion,33 interact with and modulate the functionality of IL-6,27 and serve as a nucleotide exchange factor for glucose-regulated protein 78 (GRP78), a GRP family member highly expressed in IPF while playing a pathogenic role in chronic inflammation and RA.46 Notably, hypoxia was recently shown to stimulate TNF-converting enzyme (TACE) activation, a key regulator of inflammation, via ectodomain shedding of cytokines and chemokines, a process mediated by GRP78.47 Since HYOU1, as a molecular chaperone, is mainly involved in protein synthesis, folding, and translocation of GRP78 and MMP2,29, 33, 46 the inhibition of HYOU1 in fibroblasts is anticipated to affect the viability and function of both proteins. The latter would cause a decrease in pathogenic fibroblast activation and invasion promoted by GRP78,46, 47 and in the inflammatory response mediated by MMP2, thereby reducing the secretion of various pro-inflammatory cytokines and chemokines.
Overall, our study highlights HYOU1 as a promising molecular target for the suppression of pathogenic fibroblast activation. At the same time, it provides the first-in-class small molecule HYOU1 inhibitors capable of decreasing not only the production of cytokines/chemokines but also the migratory ability and the Mmp2 expression of diseased fibroblasts. Furthermore, our research underscores the need for further development of derivatives of this series that will serve as in vivo chemical tools to probe the therapeutic role of HYOU1 in chronic inflammation, fibrosis, and cancer.
Supporting Information
Synthesis of all intermediates, final and click compounds, characterization data, NMR spectra, procedures for all in vitro, cellular and in vivo assays, and additional biological data are provided in the Supporting Information of this article. Supplementary tables showing the results of the bulk-RNA sequencing analysis (ST1, ST2) and the Ingenuity pathway analysis (ST3), as well as PDB files of complexes of 33, 48 and 52 with HYOU1 are also provided.
Author Contributions
D. P., A. N. M. and G. K. conceived the study. A. N. M. and G. K. supervised the whole study. A. N. M. designed the compounds. V. M., A. N. M. and D. G. synthesized the compounds. D. P. and F. C. performed the fibroblast and macrophage assays. D. P., V. M., F. C., M. S. and A. N. M. performed chemoproteomics studies and analysis. D. P., V. M., F. C. and A. N. M. performed target validation experiments. D. P., F. C. and C. T. performed the RNA sequencing analysis. K. D. P. and Z. F. B. conducted the cheminformatics studies. A. A. and G. S. supervised the cheminformatics studies. V. M., A. N. M., N. K. and M. C. D. performed the PK studies. N. K. and M. C. D. performed the in vivo studies. J. S. and R. L. performed the D2R/D3R dopamine receptor assays. A. N. M. and G. K. acquired funding. A. N. M., D. P. and G. K. wrote the original manuscript. D. P., V. M., A. N. M. and G. K. reviewed, and all authors edited the manuscript.
Acknowledgments
The authors acknowledge support of this work by “Research-Create-Innovate” project Drug.Art (Τ2ΕDΚ-01076) and infrastructure projects InfrafrontierGR/Phenotypos (MIS 5002135) and pMedGR (MIS 5002802) to G. K., co-financed by Greece and the European Union (European Regional Development Fund) through the Operational Program “Competitiveness, Entrepreneurship and Innovation” under NSRF 2014–2020. This work has also been supported by Horizon Europe Advanced ERC project BecomingCausal (ERC-2021-ADG, ID# 101055093) to G. K., as well as a start-up grant by the Stavros Niarchos Foundation to A. M. This work was also supported by computing time awarded on the Cyclone supercomputer of the High-Performance Computing Facility of The Cyprus Institute under preparatory project ID p114 and production project ID pr001017. The authors further acknowledge Professor Victoria Magrioti for taking the HRMS spectra of the final products, Dr. Sofia Grammenoudi for FACS sorting experiments and Dr. Vaggelis Harokopos for genomics service.
Conflict of interests
Antreas Afantitis is affiliated with NovaMechanics Ltd., a drug design company. George Kollias is founder and president of Biomedcode Hellas SA.
Open Research
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE48 partner repository with the dataset identifier PXD043742. The RNA-Seq raw data have been uploaded at the NCBI Gene Expression Omnibus49 under accession number GSE237395.