A Desaturase-Like Enzyme Catalyzes Oxazole Formation in Pseudomonas Indolyloxazole Alkaloids
Dr. Alexander O. Brachmann
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorSilke I. Probst
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorJoel Rüthi
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorDarya Dudko
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorProf. Dr. Helge B. Bode
Goethe Universität Frankfurt, Institute of Molecular Biological Science, Max-von-Laue Str. 9, 60438 Frankfurt am Main, Germany
Senckenberg Gesellschaft für Naturforschung, Senckenberganlage 25, 60325 Frankfurt am Main, Germany
Buchmann Institute for Molecular Life Sciences (BMLS), Johann Wolfgang Goethe Universität, Max-von-Laue-Straße 15, 60438 Frankfurt am Main, Germany
Max-Planck-Institute for Terrestrial Microbiology, Department of Natural Products in Organismic Interactions, 35043 Marburg, Germany
Search for more papers by this authorCorresponding Author
Prof. Dr. Jörn Piel
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorDr. Alexander O. Brachmann
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorSilke I. Probst
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorJoel Rüthi
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorDarya Dudko
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorProf. Dr. Helge B. Bode
Goethe Universität Frankfurt, Institute of Molecular Biological Science, Max-von-Laue Str. 9, 60438 Frankfurt am Main, Germany
Senckenberg Gesellschaft für Naturforschung, Senckenberganlage 25, 60325 Frankfurt am Main, Germany
Buchmann Institute for Molecular Life Sciences (BMLS), Johann Wolfgang Goethe Universität, Max-von-Laue-Straße 15, 60438 Frankfurt am Main, Germany
Max-Planck-Institute for Terrestrial Microbiology, Department of Natural Products in Organismic Interactions, 35043 Marburg, Germany
Search for more papers by this authorCorresponding Author
Prof. Dr. Jörn Piel
Eidgenössische Technische Hochschule (ETH) Zürich, Institute of Microbiology, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland
Search for more papers by this authorGraphical Abstract
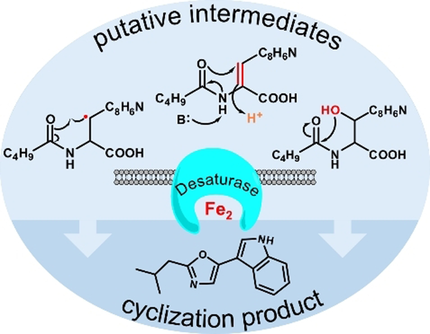
Indolyloxazole alkaloids are widespread natural products with diverse bioactivities but previously unknown biosynthesis. Transposon mutagenesis, heterologous pathway expression, and labeling experiments provide evidence for a Pseudomonas pathway involving oxazole formation by cyclization as a new reaction type catalyzed by a desaturase-like enzyme.
Abstract
Indolyloxazole alkaloids occur in diverse micro- and macroorganisms and exhibit a wide range of pharmacological activities. Despite their ubiquitous occurrence and simple structures, the biosynthetic pathway remained unknown. Here, we used transposon mutagenesis in the labradorin producer Pseudomonas entomophila to identify a cryptic biosynthetic locus encoding an N-acyltransferase and a non-heme diiron desaturase-like enzyme. Heterologous expression in E. coli demonstrates that both enzymes are sufficient to produce indolyloxazoles. Probing their function in stable-isotope feeding experiments, we provide evidence for an unusual desaturase mechanism that generates the oxazole by decarboxylative cyclization.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202014491-sup-0001-misc_information.pdf989.5 KB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1Y. Koyama, K. Yokose, L. J. Dolby, Agric. Biol. Chem. 1981, 45, 1285–1287.
- 2K. Umehara, K. Yoshida, M. Okamoto, M. Iwami, H. Tanaka, M. Kohsaka, H. Imanaka, J. Antibiot. 1984, 37, 1153–1160.
- 3T. Engl, J. Kroiss, M. Kai, T. Y. Nechitaylo, A. Svatos, M. Kaltenpoth, Proc. Natl. Acad. Sci. USA 2018, 115, E2020–E2029.
- 4J. Kroiss, M. Kaltenpoth, B. Schneider, M. G. Schwinger, C. Hertweck, R. K. Maddula, E. Strohm, A. Svatos, Nat. Chem. Biol. 2010, 6, 261–263.
- 5T. Nakashima, K. Anzai, N. Kuwahara, H. Komaki, S. Miyadoh, S. Harayama, M. D. Tianero, J. Tanaka, A. Kanamoto, K. Ando, Biol. Pharm. Bull. 2009, 32, 832–836.
- 6G. R. Pettit, J. C. Knight, D. L. Herald, R. Davenport, R. K. Pettit, B. E. Tucker, J. M. Schmidt, J. Nat. Prod. 2002, 65, 1793–1797.
- 7K. O. Hanssen, B. Schuler, A. J. Williams, T. B. Demissie, E. Hansen, J. H. Andersen, J. Svenson, K. Blinov, M. Repisky, F. Mohn, G. Meyer, J. S. Svendsen, K. Ruud, M. Elyashberg, L. Gross, M. Jaspars, J. Isaksson, Angew. Chem. Int. Ed. 2012, 51, 12238–12241; Angew. Chem. 2012, 124, 12404–12407.
- 8F. Miyake, M. Hashimoto, S. Tonsiengsom, K. Yakushijin, D. A. Horne, Tetrahedron 2010, 66, 4888–4893.
- 9I. N'Diaye, G. Guella, I. Mancini, F. Pietra, Tetrahedron Lett. 1996, 37, 3049–3050.
- 10F. Grundmann, V. Dill, A. Dowling, A. Thanwisai, E. Bode, N. Chantratita, R. ffrench-Constant, H. B. Bode, Beilstein J. Org. Chem. 2012, 8, 749–752.
- 11T. Duerfahrt, K. Eppelmann, R. Muller, M. A. Marahiel, Chem. Biol. 2004, 11, 261–271.
- 12C. T. Walsh, H. W. Chen, T. A. Keating, B. K. Hubbard, H. C. Losey, L. S. Luo, C. G. Marshall, D. A. Miller, H. M. Patel, Curr. Opin. Chem. Biol. 2001, 5, 525–534.
- 13J. O. Melby, N. J. Nard, D. A. Mitchell, Curr. Opin. Chem. Biol. 2011, 15, 369–378.
- 14Y. M. Li, J. C. Milne, L. L. Madison, R. Kolter, C. T. Walsh, Science 1996, 274, 1188–1193.
- 15Y. Zhou, A. C. Murphy, M. Samborskyy, P. Prediger, L. C. Dias, P. F. Leadlay, Chem. Biol. 2015, 22, 745–754.
- 16E. J. Rubin, B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, J. J. Mekalanos, Proc. Natl. Acad. Sci. USA 1999, 96, 1645–1650.
- 17J. I. Tietz, C. J. Schwalen, P. S. Patel, T. Maxson, P. M. Blair, H. C. Tai, U. I. Zakai, D. A. Mitchell, Nat. Chem. Biol. 2017, 13, 470–478.
- 18M. Noltemeyer, G. M. Sheldrick, H. U. Hoppe, A. Zeeck, J. Antibiot. 1982, 35, 549–555.
- 19R. Raju, O. Gromyko, V. Fedorenko, A. Luzhetskyy, R. Muller, Tetrahedron Lett. 2012, 53, 3009–3011.
- 20H. P. Bi, Y. F. Bai, T. Cai, Y. B. Zhuang, X. M. Liang, X. L. Zhang, T. Liu, Y. H. Ma, Appl. Microbiol. Biotechnol. 2013, 97, 10339–10348.
- 21H. Wang, M. G. Klein, H. Zou, W. Lane, G. Snell, I. Levin, K. Li, B. C. Sang, Nat. Struct. Mol. Biol. 2015, 22, 581–585.
- 22Y. H. Bai, J. G. McCoy, E. J. Levin, P. Sobrado, K. R. Rajashankar, B. G. Fox, M. Zhou, Nature 2015, 524, 252–256.
- 23G. Y. Zhu, M. Koszelak-Rosenblum, S. M. Connelly, M. E. Dumont, M. G. Malkowski, J. Biol. Chem. 2015, 290, 29820–29833.
- 24J. Shanklin, J. E. Guy, G. Mishra, Y. Lindqvist, J. Biol. Chem. 2009, 284, 18559–18563.
- 25J. Shanklin, E. Whittle, B. G. Fox, Biochemistry 1994, 33, 12787–12794.
- 26B. J. Wallar, J. D. Lipscomb, Chem. Rev. 1996, 96, 2625–2657.
- 27H. L. Cooper, G. Mishra, X. Huang, M. Pender-Cudlip, R. N. Austin, J. Shanklin, J. T. Groves, J. Am. Chem. Soc. 2012, 134, 20365–20375.
- 28J. Robin, M. Gueroult, R. Cheikhrouhou, M. Guicherd, V. Borsenberger, A. Marty, F. Bordes, Biotechnol. Bioeng. 2019, 116, 2451–2462.
- 29P. H. Buist, Nat. Prod. Rep. 2004, 21, 249–262.
- 30J. T. Groves, G. A. Mcclusky, R. E. White, M. J. Coon, Biochem. Biophys. Res. Commun. 1978, 81, 154–160.
- 31J. Shanklin, E. B. Cahoon, Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 611–641.
- 32D. Bím, J. Chalupský, M. Culka, E. I. Solomon, L. Rulíšek, M. Srnec, J. Am. Chem. Soc. 2020, 142, 10412–10423.
- 33R. O. Pedraza, A. Ramirez-Mata, M. L. Xiqui, B. E. Baca, FEMS Microbiol. Lett. 2004, 233, 15–21.
- 34F. Zavala, K. Takai, O. Hayaishi, J. Biol. Chem. 1983, 258, 344–351.
- 35C. Liu, A. Lei, Chem. Eur. J. 2019, 25, 7768–7770.
- 36D. Liu, S. Tang, H. Yi, C. Liu, X. Qi, Y. Lan, A. Lei, Chem. Eur. J. 2014, 20, 15605–15610.
- 37J. Petersen, K. E. Christensen, M. T. Nielsen, K. T. Mortensen, V. V. Komnatnyy, T. E. Nielsen, K. Qvortrup, ACS Comb. Sci. 2018, 20, 344–349.