Spatiotemporal Delivery of CRISPR/Cas9 Genome Editing Machinery Using Stimuli-Responsive Vehicles
Weiqi Cai
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorTianli Luo
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorProf. Lanqun Mao
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorCorresponding Author
Prof. Ming Wang
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorWeiqi Cai
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorTianli Luo
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorProf. Lanqun Mao
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorCorresponding Author
Prof. Ming Wang
Beijing National Laboratory for Molecular Science, CAS Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS), No. 2, North first street, Zhongguancun, Beijing, 100190 China
University of Chinese Academy of Sciences, No.19 (A) Yuquan Road, Shijingshan District, China
Search for more papers by this authorGraphical Abstract
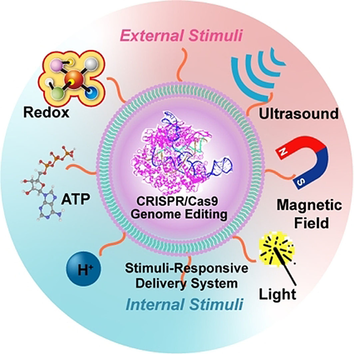
Spatiotemporal delivery of CRISPR/Cas9 genome editing machinery is of great importance for developing chemical biology tools for manipulating cellular DNA sequences to treat genetic disorders. This Minireview provides an overview of synthetic approaches and specific chemistries that have been used to develop stimuli-responsive vehicles for CRISPR/Cas9 delivery and genome editing in a controlled manner.
Abstract
Recent innovations in genome editing have enabled the precise manipulation of the genetic information of mammalians, and benefitted the development of next-generation gene therapy. Despite these advances, several barriers to the clinical translation of genome editing remain, including the intracellular delivery of genome editing machinery, and the risk of off-target editing effect. Here, we review the recent advance of spatiotemporal delivery of CRISPR/Cas9 genome editing machinery, which is composed of programmable Cas9 nuclease and a single-guide RNA (sgRNA) using stimuli-responsive nanoparticles. We discuss the specific chemistries that have been used for controlled Cas9/sgRNA delivery and intracellular release in the presence of endogenous or external signals. These methodologies can leverage biological signals found locally within disease cells, or exogenous signals administrated with spatiotemporal control, through which an improved genome editing could be achieved. We also discuss the future in exploiting these approaches for fundamental biomedical applications and therapeutic genome editing.
Conflict of interest
The authors declare no conflict of interest.
References
- 1K. A. High, M. G. Roncarolo, N. Engl. J. Med. 2019, 381, 455–464.
- 2M. H. Porteus, N. Engl. J. Med. 2019, 380, 947–959.
- 3
- 3aJ. A. Doudna, Nature 2020, 578, 229–236;
- 3bW. Cai, M. Wang, Sci. Bull. 2019, 64, 1841–1849.
- 4D. Wang, P. W. L. Tai, G. Gao, Nat. Rev. Drug Discovery 2019, 18, 358–378.
- 5H. Ledford, Nature 2020, 579, 185.
- 6C.-F. Xu, G.-J. Chen, Y.-L. Luo, Y. Zhang, G. Zhao, Z.-D. Lu, A. Czarna, Z. Gu, J. Wang, Adv. Drug Deliv. Rev. 2019 https://doi.org/10.1016/j.addr.2019.11.005.
- 7
- 7aR. Mout, M. Ray, G. Yesilbag Tonga, Y.-W. Lee, T. Tay, K. Sasaki, V. M. Rotello, ACS Nano 2017, 11, 2452–2458;
- 7bX. Zhang, B. Li, X. Luo, W. Zhao, J. Jiang, C. Zhang, M. Gao, X. Chen, Y. Dong, ACS Appl. Mater. Interfaces 2017, 9, 25481–25487;
- 7cB. Lee, K. Lee, S. Panda, R. Gonzales-Rojas, A. Chong, V. Bugay, H. M. Park, R. Brenner, N. Murthy, H. Y. Lee, Nat. Biomed. Eng. 2018, 2, 497–507;
- 7dZ. Glass, M. Lee, Y. Li, Q. Xu, Trends Biotechnol. 2018, 36, 173–185;
- 7eC. Xu, Z. Lu, Y. Luo, Y. Liu, Z. Cao, S. Shen, H. Li, J. Liu, K. Chen, Z. Chen, X. Yang, Z. Gu, J. Wang, Nat. Commun. 2018, 9, 4092;
- 7fC. Liu, T. Wan, H. Wang, S. Zhang, Y. Ping, Y. Cheng, Sci. Adv. 2019, 5, eaaw8922;
- 7gT. Wan, Y. Chen, Q. Pan, X. Xu, Y. Kang, X. Gao, F. Huang, C. Wu, Y. Ping, J. Controlled Release 2020, 246–247;
- 7hY.-W. Lee, D. C. Luther, R. Goswami, T. Jeon, V. Clark, J. Elia, S. Gopalakrishnan, V. M. Rotello, J. Am. Chem. Soc. 2020, 142, 4349–4355.
- 8
- 8aC. Liu, L. Zhang, H. Liu, K. Cheng, J. Controlled Release 2017, 266, 17–26;
- 8bN. D. Donahue, H. Acar, S. Wilhelm, Adv. Drug Delivery Rev. 2019, 143, 68–96.
- 9D. Wilbie, J. Walther, E. Mastrobattista, Acc. Chem. Res. 2019, 52, 1555–1564.
- 10L. Li, L. Song, X. Liu, X. Yang, X. Li, T. He, N. Wang, S. Yang, C. Yu, T. Yin, Y. Wen, Z. He, X. Wei, W. Su, Q. Wu, S. Yao, C. Gong, Y. Wei, ACS Nano 2017, 11, 95–111.
- 11Q. Liu, K. Zhao, C. Wang, Z. Zhang, C. Zheng, Y. Zhao, Y. Zheng, C. Liu, Y. An, L. Shi, C. Kang, Y. Liu, Adv. Sci. 2019, 6, 1801423.
- 12W. Sun, W. Ji, J. M. Hall, Q. Hu, C. Wang, C. L. Beisel, Z. Gu, Angew. Chem. Int. Ed. 2015, 54, 12029–12033; Angew. Chem. 2015, 127, 12197–12201.
- 13R. Shahbazi, G. Sghia-Hughes, J. L. Reid, S. Kubek, K. G. Haworth, O. Humbert, H.-P. Kiem, J. E. Adair, Nat. Mater. 2019, 18, 1124–1132.
- 14H. Yue, X. Zhou, M. Cheng, D. Xing, Nanoscale 2018, 10, 1063–1071.
- 15S. K. Alsaiari, S. Patil, M. Alyami, K. O. Alamoudi, F. A. Aleisa, J. S. Merzaban, M. Li, N. M. Khashab, J. Am. Chem. Soc. 2018, 140, 143–146.
- 16M. Z. Alyami, S. K. Alsaiari, Y. Li, S. S. Qutub, F. A. Aleisa, R. Sougrat, J. S. Merzaban, N. M. Khashab, J. Am. Chem. Soc. 2020, 142, 1715–1720.
- 17J. Chang, X. Chen, Z. Glass, F. Gao, L. Mao, M. Wang, Q. Xu, Acc. Chem. Res. 2019, 52, 665–675.
- 18H. Yin, C.-Q. Song, J. R. Dorkin, L. J. Zhu, Y. Li, Q. Wu, A. Park, J. Yang, S. Suresh, A. Bizhanova, A. Gupta, M. F. Bolukbasi, S. Walsh, R. L. Bogorad, G. Gao, Z. Weng, Y. Dong, V. Koteliansky, S. A. Wolfe, R. Langer, W. Xue, D. G. Anderson, Nat. Biotechnol. 2016, 34, 328–333.
- 19C. Jiang, M. Mei, B. Li, X. Zhu, W. Zu, Y. Tian, Q. Wang, Y. Guo, Y. Dong, X. Tan, Cell Res. 2017, 27, 440–443.
- 20J. B. Miller, S. Zhang, P. Kos, H. Xiong, K. Zhou, S. S. Perelman, H. Zhu, D. J. Siegwart, Angew. Chem. Int. Ed. 2017, 56, 1059–1063; Angew. Chem. 2017, 129, 1079–1083.
- 21L. Zhang, L. Wang, Y. Xie, P. Wang, S. Deng, A. Qin, J. Zhang, X. Yu, W. Zheng, X. Jiang, Angew. Chem. Int. Ed. 2019, 58, 12404–12408; Angew. Chem. 2019, 131, 12534–12538.
- 22
- 22aC. Liang, J. Chang, Y. Jiang, J. Liu, L. Mao, M. Wang, Chem. Commun. 2019, 55, 8170–8173;
- 22bQ. Tang, J. Wang, Y. Jiang, M. Zhang, J. Chang, Q. Xu, L. Mao, M. Wang, Chem. Commun. 2019, 55, 5163–5166.
- 23M. Wang, J. A. Zuris, F. Meng, H. Rees, S. Sun, P. Deng, Y. Han, X. Gao, D. Pouli, Q. Wu, Proc. Natl. Acad. Sci. USA 2016, 113, 2868–2873.
- 24J. Liu, J. Chang, Y. Jiang, X. Meng, T. Sun, L. Mao, Q. Xu, M. Wang, Adv. Mater. 2019, 31, 1902575.
- 25Q. Tang, J. Liu, Y. Jiang, M. Zhang, L. Mao, M. Wang, ACS Appl. Mater. Interfaces 2019, 11, 46585–46590.
- 26G. Chen, A. A. Abdeen, Y. Wang, P. K. Shahi, S. Robertson, R. Xie, M. Suzuki, B. R. Pattnaik, K. Saha, S. Gong, Nat. Nanotechnol. 2019, 14, 974–980.
- 27J. Liu, T. Wu, X. Lu, X. Wu, S. Liu, S. Zhao, X. Xu, B. Ding, J. Am. Chem. Soc. 2019, 141, 19032–19037.
- 28W. Zhou, H. Cui, L. Ying, X.-F. Yu, Angew. Chem. Int. Ed. 2018, 57, 10268–10272; Angew. Chem. 2018, 130, 10425–10429.
- 29R. Mo, T. Jiang, R. DiSanto, W. Tai, Z. Gu, Nat. Commun. 2014, 5, 3364.
- 30J. Deng, K. Wang, M. Wang, P. Yu, L. Mao, J. Am. Chem. Soc. 2017, 139, 5877–5882.
- 31X. Yang, Q. Tang, Y. Jiang, M. Zhang, M. Wang, L. Mao, J. Am. Chem. Soc. 2019, 141, 3782–3786.
- 32P. Wang, L. Zhang, W. Zheng, L. Cong, Z. Guo, Y. Xie, L. Wang, R. Tang, Q. Feng, Y. Hamada, K. Gonda, Z. Hu, X. Wu, X. Jiang, Angew. Chem. Int. Ed. 2018, 57, 1491–1496; Angew. Chem. 2018, 130, 1507–1512.
- 33Y. Lyu, S. He, J. Li, Y. Jiang, H. Sun, Y. Miao, K. Pu, Angew. Chem. Int. Ed. 2019, 58, 18197–18201; Angew. Chem. 2019, 131, 18365–18369.
- 34X. Chen, Y. Chen, H. Xin, T. Wan, Y. Ping, Proc. Natl. Acad. Sci. USA 2020, 117, 2395–2405.
- 35Y. Pan, J. Yang, X. Luan, X. Liu, X. Li, J. Yang, T. Huang, L. Sun, Y. Wang, Y. Lin, Y. Song, Sci. Adv. 2019, 5, eaav7199.
- 36H. Zhu, L. Zhang, S. Tong, C. M. Lee, H. Deshmukh, G. Bao, Nat. Biomed. Eng. 2019, 3, 126–136.
- 37M. Hansen-Bruhn, B. E.-F. de Ávila, M. Beltrán-Gastélum, J. Zhao, D. E. Ramírez-Herrera, P. Angsantikul, K. Vesterager Gothelf, L. Zhang, J. Wang, Angew. Chem. Int. Ed. 2018, 57, 2657–2661; Angew. Chem. 2018, 130, 2687–2691.
- 38H. Yin, K. J. Kauffman, D. G. Anderson, Nat. Rev. Drug Discovery 2017, 16, 387–399.
- 39H.-X. Wang, M. Li, C. M. Lee, S. Chakraborty, H.-W. Kim, G. Bao, K. W. Leong, Chem. Rev. 2017, 117, 9874–9906.
- 40First in vivo CRISPR candidate enters the clinic. A. Mullard, Nat. Rev. Drug Discovery 2019, 18, 656.