Steering Siglec–Sialic Acid Interactions on Living Cells using Bioorthogonal Chemistry
Christian Büll
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorTorben Heise
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorNiek van Hilten
Computational Discovery and Design Group, Centre for Molecular and Biomolecular Informatics, Radboud University Medical Center, Geert Grooteplein 26–28, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorJohan F. A. Pijnenborg
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorVictor R. L. J. Bloemendal
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorLotte Gerrits
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorEsther D. Kers-Rebel
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorDr. Tina Ritschel
Computational Discovery and Design Group, Centre for Molecular and Biomolecular Informatics, Radboud University Medical Center, Geert Grooteplein 26–28, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorDr. Martijn H. den Brok
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
Department of Anesthesiology, Pain and Palliative Medicine, Radboud University Medical Center, Geert Grooteplein 10, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorCorresponding Author
Prof. Dr. Gosse J. Adema
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorCorresponding Author
Dr. Thomas J. Boltje
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorChristian Büll
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorTorben Heise
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorNiek van Hilten
Computational Discovery and Design Group, Centre for Molecular and Biomolecular Informatics, Radboud University Medical Center, Geert Grooteplein 26–28, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorJohan F. A. Pijnenborg
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorVictor R. L. J. Bloemendal
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorLotte Gerrits
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
Search for more papers by this authorEsther D. Kers-Rebel
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorDr. Tina Ritschel
Computational Discovery and Design Group, Centre for Molecular and Biomolecular Informatics, Radboud University Medical Center, Geert Grooteplein 26–28, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorDr. Martijn H. den Brok
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
Department of Anesthesiology, Pain and Palliative Medicine, Radboud University Medical Center, Geert Grooteplein 10, 6525 GA Nijmegen, The Netherlands
Search for more papers by this authorCorresponding Author
Prof. Dr. Gosse J. Adema
Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud University Medical Center, Geert Grooteplein Zuid 32, 6525 GA Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorCorresponding Author
Dr. Thomas J. Boltje
Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
These authors contributed equally to this work.
Search for more papers by this authorGraphical Abstract
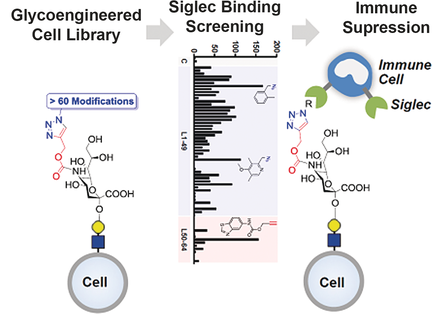
Clicking the immune system off: We report a method to rapidly reprogram the binding of sialic acid sugars on living cells to their cognate Siglec receptors through glycoengineering and click chemistry. Binding could be improved by more than 100-fold and in a selective manner. The modified cells showed potent immunosuppressive activity resulting from strong signaling through Siglecs on immune cells.
Abstract
Sialic acid sugars that terminate cell-surface glycans form the ligands for the sialic acid binding immunoglobulin-like lectin (Siglec) family, which are immunomodulatory receptors expressed by immune cells. Interactions between sialic acid and Siglecs regulate the immune system, and aberrations contribute to pathologies like autoimmunity and cancer. Sialic acid/Siglec interactions between living cells are difficult to study owing to a lack of specific tools. Here, we report a glycoengineering approach to remodel the sialic acids of living cells and their binding to Siglecs. Using bioorthogonal chemistry, a library of cells with more than sixty different sialic acid modifications was generated that showed dramatically increased binding toward the different Siglec family members. Rational design reduced cross-reactivity and led to the discovery of three selective Siglec-5/14 ligands. Furthermore, glycoengineered cells carrying sialic acid ligands for Siglec-3 dampened the activation of Siglec-3+ monocytic cells through the NF-κB and IRF pathways.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie201612193-sup-0001-misc_information.pdf3.2 MB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1
- 1aA. Varki, Glycobiology 2011, 21, 1121–1124;
- 1bB. S. Blaum, J. P. Hannan, A. P. Herbert, D. Kavanagh, D. Uhrin, T. Stehle, Nat. Chem. Biol. 2015, 11, 77–82.
- 2
- 2aR. Schauer, Curr. Opin. Struct. Biol. 2009, 19, 507–514;
- 2bA. Varki, Nature 2007, 446, 1023–1029;
- 2cP. R. Crocker, J. C. Paulson, A. Varki, Nat. Rev. Immunol. 2007, 7, 255–266.
- 3S. Pillai, I. A. Netravali, A. Cariappa, H. Mattoo, Annu. Rev. Immunol. 2012, 30, 357–392.
- 4M. S. Macauley, P. R. Crocker, J. C. Paulson, Nat. Rev. Immunol. 2014, 14, 653–666.
- 5Y. Ding, Z. Guo, Y. Liu, X. Li, Q. Zhang, X. Xu, Y. Gu, Y. Zhang, D. Zhao, X. Cao, Nat. Immunol. 2016, 17, 1167.
- 6
- 6aI. Surolia, S. P. Pirnie, V. Chellappa, K. N. Taylor, A. Cariappa, J. Moya, H. Liu, D. W. Bell, D. R. Driscoll, S. Diederichs, K. Haider, I. Netravali, S. Le, R. Elia, E. Dow, A. Lee, J. Freudenberg, P. L. De Jager, Y. Chretien, A. Varki, M. E. MacDonald, T. Gillis, T. W. Behrens, D. Bloch, D. Collier, J. Korzenik, D. K. Podolsky, D. Hafler, M. Murali, B. Sands, J. H. Stone, P. K. Gregersen, S. Pillai, Nature 2010, 466, 243–247;
- 6bT. Toubai, G. Hou, N. Mathewson, C. Liu, Y. Wang, K. Oravecz-Wilson, E. Cummings, C. Rossi, R. Evers, Y. Sun, J. Wu, S. W. Choi, D. Fang, P. Zheng, Y. Liu, P. Reddy, Blood 2014, 123, 3512–3523;
- 6cG. Y. Chen, X. Chen, S. King, K. A. Cavassani, J. Cheng, X. Zheng, H. Cao, H. Yu, J. Qu, D. Fang, W. Wu, X. F. Bai, J. Q. Liu, S. A. Woodiga, C. Chen, L. Sun, C. M. Hogaboam, S. L. Kunkel, P. Zheng, Y. Liu, Nat. Biotechnol. 2011, 29, 428–435;
- 6dG. Y. Chen, J. Tang, P. Zheng, Y. Liu, Science 2009, 323, 1722–1725;
- 6eK. Malpass, Nat. Rev. Neurol. 2013, 9, 360;
- 6fC. Büll, M. H. den Brok, G. J. Adema, Biochim. Biophys. Acta Rev. Cancer 2014, 1846, 238–246;
- 6gC. Büll, M. A. Stoel, M. H. den Brok, G. J. Adema, Cancer Res. 2014, 74, 3199–3204.
- 7
- 7aC. Büll, T. Heise, G. J. Adema, T. J. Boltje, Trends Biochem. Sci. 2016, 41, 519–531;
- 7bT. Angata, C. M. Nycholat, M. S. Macauley, Trends Pharmacol. Sci. 2015, 36, 645–660.
- 8
- 8aC. D. Rillahan, E. Schwartz, R. McBride, V. V. Fokin, J. C. Paulson, Angew. Chem. Int. Ed. 2012, 51, 11014–11018; Angew. Chem. 2012, 124, 11176–11180;
- 8bV. V. Rostovtsev, L. G. Green, V. V. Fokin, K. B. Sharpless, Angew. Chem. Int. Ed. 2002, 41, 2596–2599;
10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4 CAS PubMed Web of Science® Google ScholarAngew. Chem. 2002, 114, 2708–2711.
- 9B. Belardi, C. R. Bertozzi, Chem. Biol. 2015, 22, 983–993.
- 10J. E. Hudak, S. M. Canham, C. R. Bertozzi, Nat. Chem. Biol. 2014, 10, 69–75.
- 11
- 11aC. Büll, T. Heise, D. M. Beurskens, M. Riemersma, A. Ashikov, F. P. Rutjes, T. H. van Kuppevelt, D. J. Lefeber, M. H. den Brok, G. J. Adema, T. J. Boltje, ACS Chem. Biol. 2015, 10, 2353–2363;
- 11bN. J. Agard, C. R. Bertozzi, Acc. Chem. Res. 2009, 42, 788–797.
- 12D. C. Kennedy, C. S. McKay, M. C. Legault, D. C. Danielson, J. A. Blake, A. F. Pegoraro, A. Stolow, Z. Mester, J. P. Pezacki, J. Am. Chem. Soc. 2011, 133, 17993–18001.
- 13
- 13aS. V. Shelke, G. P. Gao, S. Mesch, H. Gathje, S. Kelm, O. Schwardt, B. Ernst, Bioorg. Med. Chem. 2007, 15, 4951–4965;
- 13bS. Kelm, J. Gerlach, R. Brossmer, C. P. Danzer, L. Nitschke, J. Exp. Med. 2002, 195, 1207–1213.
- 14M. A. Zhuravleva, K. Trandem, P. D. Sun, J. Mol. Biol. 2008, 375, 437–447.
- 15
- 15aH. Prescher, A. Schweizer, E. Kuhfeldt, L. Nitschke, R. Brossmer, ACS Chem. Biol. 2014, 9, 1444–1450;
- 15bH. H. Abdu-Allah, K. Watanabe, G. C. Completo, M. Sadagopan, K. Hayashizaki, C. Takaku, T. Tamanaka, H. Takematsu, Y. Kozutsumi, J. C. Paulson, T. Tsubata, H. Ando, H. Ishida, M. Kiso, Bioorg. Med. Chem. 2011, 19, 1966–1971;
- 15cS. Kelm, P. Madge, T. Islam, R. Bennett, H. Koliwer-Brandl, M. Waespy, M. von Itzstein, T. Haselhorst, Angew. Chem. Int. Ed. 2013, 52, 3616–3620; Angew. Chem. 2013, 125, 3704–3708.
- 16
- 16aB. Dey, R. J. Dey, L. S. Cheung, S. Pokkali, H. Guo, J. H. Lee, W. R. Bishai, Nat. Med. 2015, 21, 401–406;
- 16bK. Honda, T. Taniguchi, Nat. Rev. Immunol. 2006, 6, 644–658.
- 17
- 17aW. Chen, C. Han, B. Xie, X. Hu, Q. Yu, L. Shi, Q. Wang, D. Li, J. Wang, P. Zheng, Y. Liu, X. Cao, Cell 2013, 152, 467–478;
- 17bG. Y. Chen, N. K. Brown, W. Wu, Z. Khedri, H. Yu, X. Chen, D. van de Vlekkert, A. D'Azzo, P. Zheng, Y. Liu, elife 2014, 3, e 04066.
- 18V. C. Taylor, C. D. Buckley, M. Douglas, A. J. Cody, D. L. Simmons, S. D. Freeman, J. Biol. Chem. 1999, 274, 11505–11512.