Discrimination and Selective Enhancement of Signals in the MALDI Mass Spectrum of a Protein by Combining a Matrix-Based Label for Lysine Residues with a Neutral Matrix
David Lascoux
Protein Mass Spectrometry Laboratory, Institut de Biologie Structurale, CEA, CNRS, UJF, UMR 5075, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France, Fax: (+33) 4-3878-5494
Search for more papers by this authorDavid Paramelle
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
Search for more papers by this authorGilles Subra Dr.
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
Search for more papers by this authorMichaël Heymann
Institut de Biologie et Chimie des Protéines, Laboratoire de Bioinformatique et RMN structurales Pole BioInformatique Lyonnais, UMR 5086 CNRS, Université Lyon 1, 7 passage du Vercors, 69367 Lyon, France
Search for more papers by this authorChristophe Geourjon Dr.
Institut de Biologie et Chimie des Protéines, Laboratoire de Bioinformatique et RMN structurales Pole BioInformatique Lyonnais, UMR 5086 CNRS, Université Lyon 1, 7 passage du Vercors, 69367 Lyon, France
Search for more papers by this authorJean Martinez Prof.
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
These authors contributed equally to this research.
Search for more papers by this authorEric Forest Dr.
Protein Mass Spectrometry Laboratory, Institut de Biologie Structurale, CEA, CNRS, UJF, UMR 5075, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France, Fax: (+33) 4-3878-5494
These authors contributed equally to this research.
Search for more papers by this authorDavid Lascoux
Protein Mass Spectrometry Laboratory, Institut de Biologie Structurale, CEA, CNRS, UJF, UMR 5075, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France, Fax: (+33) 4-3878-5494
Search for more papers by this authorDavid Paramelle
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
Search for more papers by this authorGilles Subra Dr.
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
Search for more papers by this authorMichaël Heymann
Institut de Biologie et Chimie des Protéines, Laboratoire de Bioinformatique et RMN structurales Pole BioInformatique Lyonnais, UMR 5086 CNRS, Université Lyon 1, 7 passage du Vercors, 69367 Lyon, France
Search for more papers by this authorChristophe Geourjon Dr.
Institut de Biologie et Chimie des Protéines, Laboratoire de Bioinformatique et RMN structurales Pole BioInformatique Lyonnais, UMR 5086 CNRS, Université Lyon 1, 7 passage du Vercors, 69367 Lyon, France
Search for more papers by this authorJean Martinez Prof.
Institut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, Universités Montpellier 1 et 2, 15 avenue Charles Flahault, 34000 Montpellier, France, Fax: (+33) 4-6754-8654
These authors contributed equally to this research.
Search for more papers by this authorEric Forest Dr.
Protein Mass Spectrometry Laboratory, Institut de Biologie Structurale, CEA, CNRS, UJF, UMR 5075, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France, Fax: (+33) 4-3878-5494
These authors contributed equally to this research.
Search for more papers by this authorGraphical Abstract
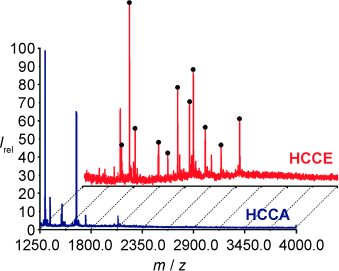
Unterscheidung gelungen: Der N-Hydroxysuccinimidester von HCCA (α-Cyan-4-hydroxyzimtsäure) wurde als Markierungsmittel genutzt, um das MALDI-MS-Signal bestimmter, gering konzentrierter Peptide in einer aus Cytochrom c erhaltenen proteolytischen Mischung relativ zu den Signalen anderer Peptide zu verstärken. Der erwünschte Effekt trat nur bei der neutralen MALDI-Matrix HCCE (dem Methylester von HCCA) auf (siehe Massenspektren).
Supporting Information
Supporting information for this article is available on the WWW under http://www.wiley-vch.de/contents/jc_2001/2007/z700811_s.pdf or from the author.
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1J. Borch, T. J. Jorgensen, P. Roepstorff, Curr. Opin. Chem. Biol. 2005, 9, 509–516.
- 2T. E. Wales, J. R. Engen, Mass Spectrom. Rev. 2006, 25, 158–170.
- 3E. O. Hochleitner, M. K. Gorny, S. Zolla-Pazner, K. B. Tomer, J. Immunol. 2000, 164, 4156–4161.
- 4S. D. Maleknia, C. Y. Ralston, M. D. Brenowitz, K. M. Downard, M. R. Chance, Anal. Biochem. 2001, 289, 103–115.
- 5P. Novak, G. H. Kruppa, M. M. Young, J. Schoeniger, J. Mass Spectrom. 2004, 39, 322–328.
- 6M. Trester-Zedlitz, K. Kamada, S. K. Burley, D. Fenyo, B. T. Chait, T. W. Muir, J. Am. Chem. Soc. 2003, 125, 2416–2425.
- 7S. C. Alley, F. T. Ishmael, A. D. Jones, S. J. Benkovic, J. Am. Chem. Soc. 2000, 122, 6126–6127.
- 8K. M. Pearson, L. K. Pannell, H. M. Fales, Rapid Commun. Mass Spectrom. 2002, 16, 149–159.
- 9D. R. Müller, P. Schindler, H. Towbin, U. Wirth, H. Voshol, S. Hoving, M. O. Steinmetz, Anal. Chem. 2001, 73, 1927–1934.
- 10A. Pashkova, E. Moskovets, B. L. Karger, Anal. Chem. 2004, 76, 4550–4557.
- 11M. Karas, M. Glückmann, J. Schäfer, J. Mass Spectrom. 2000, 35, 1–12.
10.1002/(SICI)1096-9888(200001)35:1<1::AID-JMS904>3.0.CO;2-0 CAS PubMed Web of Science® Google Scholar
- 12R. Knochenmuss, Analyst 2006, 131, 966–986.
- 13G. H. Dihazi, A. Sinz, Rapid Commun. Mass Spectrom. 2003, 17, 2005–2014.
- 14B. Schilling, R. H. Row, B. W. Gibson, X. Guo, M. M. Young, J. Am. Soc. Mass Spectrom. 2003, 14, 834–850.
Citing Literature
This is the
German version
of Angewandte Chemie.
Note for articles published since 1962:
Do not cite this version alone.
Take me to the International Edition version with citable page numbers, DOI, and citation export.
We apologize for the inconvenience.