Detecting New Lesions Using a Large Language Model: Applications in Real-World Multiple Sclerosis Datasets
Abstract
Objective
Neuroimaging is routinely utilized to identify new inflammatory activity in multiple sclerosis (MS). A large language model to classify narrative magnetic resonance imaging reports in the electronic health record (EHR) as discrete data could provide significant benefits for MS research. The objectives of the current study were to develop such a prompt and to illustrate its research applications through a common clinical scenario: monitoring response to B-cell depleting therapy (BCDT).
Methods
An institutional ecosystem that securely connects healthcare data with ChatGPT4 was applied to clinical MS magnetic resonance imaging reports in a single institutional EHR (2000–2022). A prompt (msLesionprompt) was developed and iteratively refined to classify the presence or absence of new T2-weighted lesions (newT2w) and contrast-enhancing lesions (CEL). The multistep validation included evaluating efficiency (time and cost), comparison with manually annotated reports using standard confusion matrix, and application to identifying predictors of newT2w/CEL after BCDT start.
Results
Accuracy of msLesionprompt was high for detection of newT2w (97%) and CEL (96.8%). All 14,888 available reports were categorized in 4.13 hours ($28); 79% showed no newT2w or CEL. Data extracted showed expected suppression of new activity by BCDT (>97% monitoring magnetic resonance images after an initial “rebaseline” scan). Neighborhood poverty (Area Deprivation Index) was identified as a predictor of inflammatory activity (newT2w: OR 1.69, 95% CI 1.10–2.59, p = 0.017; CEL: OR 1.54, 95% CI 1.01–2.34, p = 0.046).
Interpretation
Extracting discrete information from narrative imaging reports using an large language model is feasible and efficient. This approach could augment many real-world analyses of MS disease evolution and treatment response. ANN NEUROL 2025;98:308–316
Among the many applications of artificial intelligence (AI)-enabled large language models (LLMs) in medicine,1, 2 is their ability to derive discrete metrics from the massive amounts of narrative data in electronic health records (EHRs). This can include deriving indices of disease severity from narrative notes and reports, or of episodic events such as phases of oncologic treatment.2
For individuals with multiple sclerosis (MS), a chronic autoimmune neurological disease, there is widespread use of neuroimaging to monitor for episodic inflammatory activity. Radiology reports describe the presence of new T2-weighted lesions (newT2w) and of active, contrast-enhancing lesions (CEL) on magnetic resonance imaging (MRI), often using a structured narrative, but the key metrics (presence or absence of lesions) do not exist as discrete data for analysis within the EHR. AI-enabled analysis of narrative MRI reports to extract such data could support clinical research by expediting the ability to study patterns and predictors of new inflammatory activity in real-world clinical cohorts.
Previous research has shown the broad potential of LLMs across medical contexts, including for MS care and research.3 For example, to improve the efficiency of clinical care, LLMs can be utilized to respond to common questions from patients with MS, being perceived at times as even more empathetic than neurologist responses,4 or to automate transcription and ordering during clinical visits, as is being piloted using AI-enabled medical scribes. For MS research, the ability of LLMs to extract information about a patient's disability status from medical notes is being evaluated utilizing commercially available MS registries.5 In the radiological context of MS, there is emerging use of AI-enabled quantification of new inflammatory activity and brain volume loss from clinical images,6 including using Chat Generative Pre-Trained Transformer 4 (ChatGPT4)-vision.7 Given the prevalence of narrative MRI reports in standard EHRs, the ability to extract discrete data fields from these reports could substantially augment and accelerate the conduct of real-world research in MS.
The current study aimed to iteratively develop, refine, and clinically validate such an AI-enabled prompt. For this, Versa, an AI ecosystem that connects ChatGPT4 with healthcare data securely and enables the use of this technology for research using institutional clinical records, was utilized. After development, iterative refinement, and validation of the LLM prompt, its utility and validity in clinical research were illustrated in three ways. First, the efficiency of the prompt in analyzing all institutionally available MRI reports was queried. Then, the prompt was applied to a common, real-world clinical scenario: monitoring of MRI activity after initiation of B-cell depleting therapy. In their clinical development (ocrelizumab,8 ublituximab,9 ofatumumab10) and clinical experience (rituximab11), these medications near-abrogated CEL, and dramatically reduced newT2w, particularly after the first 6-month initiation period. The prompt was validated in this cohort, then the extracted data were used to ask a clinical question: which factors predict treatment response? Altogether, these analyses are intended to demonstrate the efficiency and impact of LLM models in leveraging historical narrative medical records for research.
Methods
Study Design and Setting
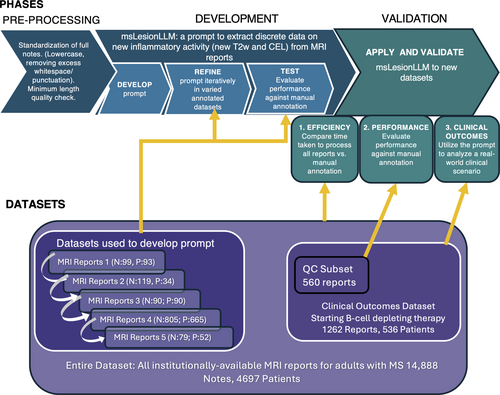
In this single-center real-world study, an LLM-enabled “prompt” to extract information about new MS inflammatory activity from brain MRI reports (msLesionprompt) was created and refined through a multistage iterative process, then validated in a multistep process shown in Figure 1. Clinical radiology reports from brain MRIs for adults with MS receiving care at the UCSF MS Center, a large and diverse tertiary care center in Northern California (USA), were utilized.12, 13 From this electronic health record, a modified algorithm to identify MS patients through the UCSF Electronic Health Record (EHR: Epic)14 was used. All MRI reports available for individuals with MS from the historical EHR were extracted, yielding 14,888 MRI reports (N = 4,697 patients) generated from a pool of 525 radiologist readers (attending and trainee radiologists) from 2000 to 2022. From these MRI reports, specific datasets were utilized for each phase of prompt development and validation, as shown in Figure 1 and detailed below.
Ethical Approvals
The UCSF Committee of Human Research approved the study protocol for retrospective analysis of EHR-derived MS data with no patient contact (Ref #13–11,686).
Phase I. MsLesionprompt Development
LLM Program: Versa LLM
The Versa application programming interface was used, which is the Microsoft Azure OpenAI generative pretrained transformer (GPT) 4 4 [GPT4o, specific model = gpt-4o-2024-05-13] application programming interface that is protected health information- and intellectual property-compliant at the University of California, San Francisco (UCSF), as described in detail.15 Versa GPT-4 has a token limit of 8,192 tokens, defined as the unit used to compute text length.16
Iterative MsLesionprompt Engineering Strategy
The development of the new prompt commenced by drawing inspiration from analogous zero shot programming projects and refined in accordance with OpenAI's best practices for prompt design. The principal refinement technique involved requesting outputs adhering to the RFC8259 JSON standard, accompanied by explanatory feedback pinpointing the textual evidence from the notes influencing the determination. For both newT2 and CEL, the msLesionprompt outputs a “1” (presence), “0” (absence), or “XX” (not found/a report that is incomplete or empty). The prompt was applied to each of the 5 datasets sequentially and refined iteratively (as shown in Supplementary Table S1). Each dataset was a convenience dataset, and had been previously manually annotated for newT2w and CEL by a neurologist (R.B.) based on the radiologist's report, for clinical research projects evaluating MRI outcomes. Datasets evaluated inflammatory activity associated with contraceptives in young women17 and with postpartum relapses18 (enriched for new MRI activity), with transition from S1P receptor modulators to B-cell depleting therapy (lower activity19), and included an unpublished research project on MS DMTs. Notes pre-processing steps included standardization of full notes (lowercase, removing excess whitespace/punctuation), and ensuring they met minimum length quality check. For each dataset, the LLM output was compared with manual annotations for both newT2w and CEL (Fig 1). Subsequent revisions of the prompt incorporated this feedback to enhance the specificity with which the target description was articulated. In an additional initial engineering step, the prompt was run through 103 manually annotated MRI reports where the header indicated that gadolinium-based contrast agent had not been performed; it accurately generated a “–1” for 94 out of 94 reports indicating that no contrast had been given, and accurately generated a “0” or “1” for 9 out of 9 of these 103 reports where the header as stored in the EHR indicated “without contrast,” but the report itself indicated that contrast had been given and that the radiologist had reviewed these sequences. This step ensured that the prompt could accurately determine where or not contrast had been given.
Final MsLesionprompt Performance
The first and final prompts engineered are presented in Supplementary Table S1. MRI impression text from cohort-identified patients is ingested directly from the EHR into the MS datamart pipeline describe above.14 These notes are then automatically downloaded, pre-processed, formatted into prompts, and submitted to the LLM API. Finally, the LLM responses are automatically saved and parsed into structured columns for analysis and review. This end-to-end automation minimizes potential human error and significantly reduces processing time. The categorical prompt output (1 = presence, 0 = absence, −1 = uninterpretable) was compared with the categorical manual annotation for each outcome (newT2w, CEL); the low number of uninterpretable reports were ommitted from the standard confusion matrix generated. To ensure that the prompt was robust to fluctuations in the LLM, the notes were then processed in triplicate and the results across the three trials compared.
Phase II. MsLesionprompt Validation
Validation 1: Efficiency of LLM-enabled data extraction versus manual note annotation at scale. Dataset utilized. For this step, all MRI reports available for individuals with MS (defined above) from the historical EHR were extracted, yielding 14,888 MRI reports (N = 4,697 patients) generated from a pool of 525 radiologist readers (attending and trainee radiologists) from 2000 to 2022. The Versa LLM prompt was applied to each of these. The time required and output generated were tabulated. For comparison to manual annotation, the time required for a neurologist with MS expertise (R.B.) to review and annotate a subset of N = 100 notes was measured, as well as for two neurologist trainees (one fellow, one resident; N = 20 notes).
Validations 2 and 3: Overview of Datasets Utilized
A dataset was generated from the total MRI reports available, for all adults with an MS diagnosis initiating antiCD20 therapy who had at least 1 post-initiation contrast-enhanced MRI and reviewed within our institution, and at least 1 pre-initiation MRI performed for comparison (n = 536 patients, 1,262 MRIs). Further details of categorization of MRI reports for inclusion are provided in Supplementary Figure S1. The LLM prompt was applied to reports for all MRIs obtained while patients were on antiCD20 therapy.
Validation 2. Validation of LLM-enabled data extraction vs. msLesionprompt in an independent dataset. The categorical prompt output was compared with the categorical manual annotation for each outcome (newT2w, CEL) for a subset of 560 of the 1,262 MRI reports: all MRI reports categorized as “positive” for either new T2w or CEL, and randomly selected reports categorized as “negative” for CEL (325 reports) and “negative” for new T2w (250 reports). A standard confusion matrix was generated.
Validation 3: Application of msLesionprompt to clinical research, using a real-world clinical scenario as a use case. Prompt output was then analyzed for the entire set of 1,262 MRI reports.
Presence of inflammatory activity on MRI after antiCD20 start. The presence of newT2w and CEL after antiCD20 start was categorized for all MRIs, then summarized according to specific timepoints: within or beyond 6 months of antiCD20 start, first MRI after 6 months on antiCD20 therapy (“rebaseline”), and first MRI after the “rebaseline” MRI (“surveillance”).
Predictors of inflammatory activity after antiCD20 start. Clinical and demographic covariates were extracted from the EHR using a modified previously described pipeline,14 and categorized for the current analyses as follows. Demographic variables were: sex, age, body mass index (in kg/m2) at MRI, race, ethnicity, and Area Deprivation Index (ADI). Race and ethnicity were further categorized as: white non-Hispanic/Asian/Pacific Islander, Hispanic/black, and other/decline/unknown, as prior work identified these as social constructs relevant to understanding disparities in our clinical MS population.12, 20 The ADI was calculated by aggregating publicly available data from the Census and American Community Survey utilizing the uszipcode21 and census22 python packages, and calculated as previously described.23 Clinical variables were: MS duration since symptom onset, disease severity (Expanded Disability Status Scale24), and MS type: relapsing (relapsing–remitting MS, clinically isolated syndrome, progressive–relapsing MS), progressive non-relapsing (primary progressive MS, secondary progressive MS) and clinically silent (radiologically isolated syndrome).
Statistical Analysis
To compare the performance of the AI-enabled LLM prompt classification of inflammatory activity from MRI reports against neurologist-annotated reports, a standard confusion matrix was applied for each outcome (newT2w, CEL). To determine whether patient characteristics influenced agreement, we examined the association between agreement and patient demographic and clinical characteristics using logistic regression and χ2 tests as appropriate for data types. To compare the efficiency of LLM over manual review of MRI reports, the average number of seconds per scan was calculated (for neurologists and neurology trainees) and then imputed for the entire dataset of available MRI reports. These durations were compared using t tests. To characterize the presence and tempo of active MRIs, the percentage of MRIs demonstrated newT2w and CEL were calculated at each timepoint. Finally, to understand the contribution of demographic or clinical factors to having persistently active MRIs after initiation of B=cell depleting therapy, nominal logistic regressions were utilized. Factors that had a p value of <0.10 in univariable associations for either MRI outcome at each timepoint were included in a multivariable regression. The JMP 17 Pro SAS Package (Cary, NC, USA) was used.25
Results
Phase I: MsLesionprompt Performance
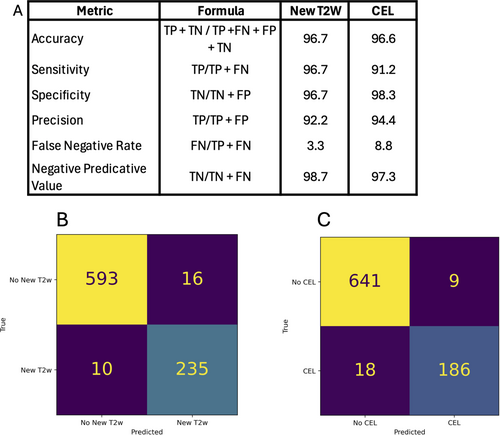
In the final dataset utilized for the prompt development (n = 850 reports; 6 were uninterpretable and omitted; 635 patients), for the detection of new T2w lesions, accuracy was 97.5%, sensitivity was 97.4%, specificity was 95.9%, and negative predictive value was 93.6%. For the detection of CEL, accuracy was 97.0%, sensitivity was 98.6%, specificity was 91.2%, and negative predictive value was 95.4% (Fig 2). When the prompt was run in triplicate on the 856 reports, accuracy across the three trials was 98.2%.
Phase II: Multistep Validation
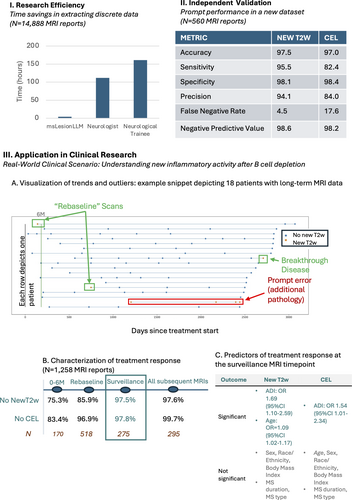
Validation 1: Efficiency of msLesionprompt versus manual note annotation. The time taken for msLesionprompt to categorize all 14,888 MRIs using the institutionally-secure GPT-4o was 4.13 hours. The pre-processing step itself took 6,200 ms for this dataset (<1 minute). The cost was $28 (for 9,440,370 tokens). Overall, 7% of all MRI reports available were categorized as showing newT2w and CEL, 9% newT2w alone, 4% CEL, and 79% no newT2w or CEL.
By comparison, at a neurologist's measured annotation rate of 27 seconds per report, reading these reports would have taken 111.7 hours (27-fold longer). This timing assumes no breaks, including cognitive breaks. This corresponds to 2.8 40-hour work weeks. In comparison with neurology trainees, with a measured rate of 39 seconds per MRI report, this would have taken 161 hours (40-fold longer) and 4 work weeks (Fig 3, Panel I).
Overview of Datasets for Clinical Validation 2 and 3
Clinical Overview
Altogether, 1,262 MRI reports from 536 individuals with MS were included in the analysis. This was a diverse cohort of individuals, with a median age of 40.4 years (IQR 33.4–51.1 years), MS duration 4.8 years (IQR 1.4–11.4 years), and median Expanded Disability Status Scale 2.5 (IQR 1.5–4); 57% identified as non-Hispanic white (15.7% Hispanic, 8.2% Asian/Pacific Islander, 7.6% black, 11.7% other/decline/unknown). Most participants (69.8%) initiated ocrelizumab. Full demographic and clinical characteristics are summarized in Supplementary Table S2.
Validation 2: Performance of msLesionprompt in an independent dataset. In the subset of 560 MRI reports reviewed, when compared with manual annotations, the prompt output had an accuracy of 97.5% for newT2w and 97.0% for CEL (Fig 3, Panel II). Examples of both simple and complex MRI reports accurately classified by the prompt, as well as of MRI reports misclassified for various reasons, are provided in Supplementary Figure S2. There was no association between patient age, sex, race, ethnicity, or MS type or duration and agreement between the prompt output and manual annotation (p > 0.05 for each comparison).
Validation 3: Utility in clinical research: A real-world clinical scenario use case. The prevalence of inflammatory activity after antiCD20 therapy start. The LLM output applied to longitudinal MRI results was graphically shown (Fig 3, Panel III A), enabling rapid visual inspection of longitudinal MRIs to identify specific outliers or groups of patients. Examples include individuals who experienced disease breakthrough, as well as individuals who had non-MS-related abnormalities misclassified by the prompt.
In the entire dataset described above (1,262 interpretable MRI reports, 533 individuals), 95.9% of MRIs showed no CEL, and 89.7% showed no newT2w after antiCD20 therapy start. When analyzing MRI timepoints according to current practice, at the “rebaselining” MRI at least 6 months after treatment start, 96.9% of reports described no CEL, and 85.9% no newT2w. At the first “surveillance” comparison MRI against this rebaselined MRI, 97.8% of MRIs showed no CEL, and 97.4% showed no newT2w. After this timepoint, in a total of 297 MRIs, 1 (0.3%) showed CEL and 7 (2.4%) showed newT2w. Further timepoints are summarized in Supplementary Table 4.
Predictors of ongoing inflammatory activity after antiCD20 therapy start. At the “rebaseline” scan, no covariates evaluated (age, sex, race, body mass index, MS type of duration) met the threshold significance in multivariable associations for either MRI outcome (newT2w or CEL). At the subsequent “surveillance” scan, higher ADI was in multivariable analyses associated with increased OR for both new T2w (OR 1.69, 95% CI 1.10–2.59, p = 0.017) and CEL (OR 1.54, 95% CI 1.01–2.34, p = 0.046). Unexpectedly, older age was also associated with newT2w at this timepoint (OR 1.09, 95% CI 1.02–1.17, p = 0.011; Fig 3, Panel III C, full details in Supplementary Table S5).
Discussion
The current analyses describe the development of AI-enabled LLM to securely, efficiently, and accurately extract meaningful discrete metrics from narrative reports within an institutional secure framework. In the multipronged validation efforts, the msLesionprompt was orders of magnitude more efficient than manual annotation. Furthermore, its potential impact on clinical research was shown by applying it to a common real-world clinical scenario (B-cell depleting therapy treatment outcomes in a large diverse MS cohort), where it recapitulated treatment efficacy, as observed in pivotal clinical trials.
Recent studies have shown the broad potential of LLMs in medicine, including in the MS examples described above.1 In radiological contexts, LLMs have shown great potential to analyze and explain reports. For example, they have been used to omit complex medical jargon and improve patient readability of complex reports—such as foot and ankle imaging reports,26 and other imaging modalities and body regions.27 LLMs have also been specifically used to analyze brain MRI reports. One study utilized a cohort of 2,398 radiological reports, in 595 of which the LLM identified the presence of a headache based on the provided clinical context. It was able to analyze the MRI findings of these reports, and determine whether such findings were normal or abnormal with 96.0% sensitivity and 98.9% specificity.28 Although many hospitals and universities have utilized LLMs for imaging data, few have applied them in clinical settings.29
The current msLesionprompt was rapidly and accurately able to classify from a large number of reports, including reports requiring sophisticated interpretation, such as the presence of other non-MS related pathology. The limitations of LLM application to clinical and radiological settings in the current study echo those in the broader literature. In radiology, performance has been noted to vary across different imaging modalities, such as X-rays, compared with computed tomography and MRI reports.26 Additionally, there are limitations in clinical correlations. For example, when tasked with inferring causality between MRI findings and headaches, an LLM showed a sensitivity of 88.2% and a specificity of 73%, highlighting challenges in correlating MRI abnormalities with patient-reported headaches.28 Furthermore, multiple studies have cautioned that findings can be biased depending on the data the AI is trained on. This includes using human reports as the “gold standard,”15 basing readability scores on specific scales,27 or using outdated data for training.30 Indeed, when evaluating the msLesionprompt performance, several errors in neurologist interpretation were identified, which points to the need for a manual revision in cases of disagreement—as both the prompt and the “gold standard” radiologist's report and clinician's annotations are subject to error.
Although automated interpretation of scans themselves would represent an ultimate application of AI, the current analyses permit the rapid generation of a large dataset of discrete data that can be utilized for other clinical research applications. Examples could include comparison of treatment response with various medications, and identification of predictors, analysis of healthcare utilization and costs associated with new radiological inflammatory activity, comparison of disease severity across demographic groups or treatment epochs, or identification of an EHR-based signature of prodromal inflammatory activity.
When applied to the real-world clinical scenario, the msLesionprompt showed value, including the ability to rapidly and accurately generate discrete data from narrative notes, saving weeks of tedious and painstaking human effort. The main clinical research findings, near-abrogation of CEL and newT2w after the first 6 months on B-cell depleting therapy, are consistent with those from the products' clinical development,8-10 and permit several scientific insights and takeaways. For example, the value of repeated imaging after a first negative “rebaselining” evaluation, as thereafter >97% MRIs show no new inflammatory activity at all. Furthermore, neighborhood deprivation (ADI) was associated with ongoing inflammatory activity, in line with other studies pointing to social determinants of health as predictors of MS course and potentially treatment response.31-33 Mechanisms for this potential association require further research. Furthermore, the unexpected association between older age and elevated odds of newT2 lesions after antiCD20 therapy start raises the possibility that new microvascular lesions associated with advancing age are mis-characterized by radiologists as new MS lesions. Even so, caution must be taken when drawing conclusions from this current real-world dataset, which was intended as a use case application of msLesionprompt and not an exhaustive exploration of the efficacy of antiCD20 therapy (discussed fully in Supplementary Table S6). Examples of limitations include individual patient/clinical decision-making dictating the type and frequency of surveillance MRI, as well as access factors (geographic, payor) dictating timing and site (institutional vs external) of clinical MRI acquisition. Finally, the msLesionprompt scalability should be explored with validation in other health systems, independent datasets and linguistic settings; it could be limited in settings where radiologists do not explicitly compare images with prior scans in their reports.
The current study showed the feasibility, efficiency, and scientific utility of developing a protected health information-compliant prompt to rapidly detect new inflammatory activity in individuals with MS. Further clinical application and validation of the current msLesionprompt can include its use for rapid automated determination of inflammatory activity from MRI reports, to support large-scale observational research across health systems in MS, including demographic or clinical predictors of treatment response. This would need to occur in parallel with the governance structures that ensure the safe and ethical use of generative artificial intelligence tools both within our institution, which is one of the few to deploy protected health information-compliant LLMs, and beyond; with clarification of their use as medical devices, as well as with the development of better strategies to combat algorithmic bias in medicine.15
Acknowledgements
We thank the research participants who made this study possible. This research study was funded by F. Hoffmann-La Roche as part of Integrative Neuroscience Collaborations Network.
Author Contributions
S.P., K.K., J.W., V.S., R.H., and R.B. contributed to the conception and design of the study; S.P., K.K., N.S., K.H., J.W., D.P., C.C., W.R., J.W., V.S., R.H., R.B., and A.A. contributed to the acquisition and analysis of data; and S.P., N.S., A.A., J.J., J.W., V.S., R.H., and R.B. drafted a significant portion of the manuscript or figures.
Potential Conflicts of Interest
S.P., K.K., N.S., K.H., J.W., D.P., C.C., W.R., A.A., and A.R.: nothing to report. J.W. and V.S. are employees of F. Hoffmann-La Roche, Basel, Switzerland. R.H. has received personal compensation for serving on a Scientific Advisory or Data Safety Monitoring board for Novartis and Roche, as well as research funding from Roche/Genentech, which make B-cell depleting therapies. R.B. receives research support from Roche Genentech and Novartis, and has received personal consulting fees from TG Therapeutics, which make B-cell depleting therapies.
Open Research
Data Availability
Deidentified, aggregate data will be made available upon reasonable request to qualified investigators with approval by the authors. Requests can be made to the corresponding author by email.




