Ultrasensitive NGS MRD assessment in Ph+ ALL: Prognostic impact and correlation with RT-PCR for BCR::ABL1
Nicholas J. Short and Elias Jabbour equally contributed to this work.
Abstract
Reverse transcription polymerase chain reaction (RT-PCR) for BCR::ABL1 is the most common and widely accepted method of measurable residual disease (MRD) assessment in Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL); however, RT-PCR may not be an optimal measure of MRD in many cases of Ph+ ALL. We evaluated the clinical impact of a highly sensitive next-generation sequencing (NGS) MRD assay (sensitivity of 10−6) and its correlation with RT-PCR for BCR::ABL1 in patients with Ph+ ALL. Overall, 32% of patients had a discordance between MRD assessment by RT-PCR and NGS, and 31% of patients who achieved NGS MRD negativity were PCR+ at the same timepoint. Among eight patients with long-term detectable BCR::ABL1 by PCR, six were PCR+/NGS−. These patients generally had stable PCR levels that persisted despite therapeutic interventions, and none subsequently relapsed; in contrast, patients who were PCR+/NGS+ had more variable PCR values that responded to therapeutic intervention. In a separate cohort of prospectively collected clinical samples, 11 of 65 patients (17%) with Ph+ ALL who achieved NGS MRD negativity had detectable BCR::ABL1 by PCR, and none of these patients relapsed. Relapse-free survival and overall survival were similar in patients who were PCR+/NGS− and PCR−/NGS−, suggesting that PCR for BCR::ABL1 did not provide additional prognostic information in patients who achieved NGS MRD negativity. NGS-based assessment of MRD is prognostic in Ph+ ALL and identifies patients with low-level detectable BCR::ABL1 who are unlikely to relapse nor to benefit from therapeutic interventions.
1 INTRODUCTION
Detection of measurable residual disease (MRD) is highly prognostic for relapse and long-term survival in acute lymphoblastic leukemia (ALL).1 Reverse transcription polymerase chain reaction (RT-PCR) for BCR::ABL1 is the recommended method of MRD assessment for Philadelphia chromosome-positive (Ph+) ALL in some consensus guidelines.2 Achievement of a complete molecular response (CMR), usually defined as absence of detectable BCR::ABL1 transcripts by RT-PCR, is an important response endpoint that is associated with superior outcomes in patients with Ph+ ALL undergoing frontline therapy.3 Failure to achieve a CMR is associated with high rates of relapse and may be an indication for MRD-directed approaches such as blinatumomab or allogeneic stem cell transplantation (alloSCT).2, 4 However, RT-PCR for BCR::ABL1 is an imperfect MRD method for predicting risk of relapse in Ph+ ALL, as 20–30% of patients will relapse despite achieving CMR, and some patients may have long-term survival despite persistently detectable BCR::ABL1 transcripts.3, 5
RT-PCR for BCR::ABL1 has poor correlation with some other MRD methods, including flow cytometry and RT-PCR for immunoglobin (IG) or T-cell receptor (TR) rearrangements, despite similar sensitivities of these assays.5-8 In one study, nearly one-quarter of patients had detectable BCR::ABL1 by RT-PCR but were MRD negative by PCR for IG/TR.8 Cell sorting assays in these discordant cases suggest the BCR::ABL1 transcripts were present in nonblast population, including in cells of myeloid lineage, thus identifying a population of patients with “chronic myeloid leukemia (CML)-like biology”. In these patients, RT-PCR for BCR::ABL1 may not be optimal for MRD assessment, as the BCR::ABL1 fusion may be present in multiple nonleukemic cell lineages and therefore may not represent residual leukemia when detected.5
Next-generation sequencing (NGS)-based MRD assessment also tracks clonal IG/TR rearrangements but has superior sensitivity to these RT-PCR assays.2 Ultrasensitive NGS assays can detect MRD with an analytical sensitivity of 10−6 and further risk stratify patients with negative or indeterminate MRD by multiparameter flow cytometry or PCR for IG/TR.9-11 Several studies across a variety of clinical contexts have suggested that even low levels of MRD between 10−4 and 10−6 detected by NGS impact risk of relapse.9, 12, 13 However, despite its established prognostic utility in Philadelphia chromosome-negative ALL and some other lymphoid diseases,9-12, 14-16 the clinical impact of NGS-based MRD has not been systematically evaluated in patients with Ph+ ALL. In this retrospective analysis, we sought to correlate MRD detected by RT-PCR for BCR::ABL1 and by NGS for IG/TR and to determine the prognostic impact of NGS MRD.
2 METHODS
2.1 Study design and participants
This is a retrospective study evaluating the prognostic impact of a highly sensitivity NGS MRD assay in patients with Ph+ ALL. Eligible patients received frontline ALL therapy at our institution between March 2006 and June 2019 using a backbone of hyper-CVAD-based therapy plus a BCR::ABL1 TKI and achieved complete remission as best response. Patients who received frontline blinatumomab-based therapies were excluded. In the primary analysis, patients were selected for inclusion based on the availability of a banked pretreatment bone marrow sample (for purposes of clonality determination) and at least 1 post-treatment remission bone marrow sample that met one or more of three prespecified criteria: (1) remission marrow collected between 1 month and 6 months from the start of frontline treatment, regardless of MRD response by RT-PCR for BCR::ABL1 (“early MRD” group), (2) remission marrow collected >2 years from the start of treatment in a patient who had been in CMR for at least 1 year (“long-term PCR−” group), and (3) remission marrow collected >2 years from the start of treatment in a patient who had persistently detectable, stable PCR values for at least 1 year (“long-term PCR+” group). For criteria 2–3, patients were required to be in continuous first remission without prior alloSCT. This study was conducted at a single academic center (The University of Texas MD Anderson Cancer Center [UTMDACC]). This study was approved by the Institutional Review Board of UTMDACC and was conducted in accordance with the Declaration of Helsinki.
2.2 BCR::ABL1 RT-PCR for MRD assessment
MRD assessment was performed in our clinical laboratory on fresh bone marrow samples or peripheral blood, as previously described.17 The sensitivity of this assay is between 0.01% (10−4) and 0.001% (10−5). Per our institutional practice, RT-PCR for BCR::ABL1 for MRD was assessed with each remission bone marrow, approximately every 1–3 months for the first year of treatment, then approximately every 3–6 months thereafter. RT-PCR MRD information was available from the clinical laboratory and was available to clinicians for decision-making.
2.3 NGS assay for MRD assessment
The clonoSEQ MRD assay (Adaptive Biotechnologies Co., Seattle, WA) is an NGS-based immunosequencing assay that was used for this analysis, as previously described.9 Briefly, pretreatment genomic DNA derived from stored bone marrow specimens was sequenced using multiplex PCR assays composed of primers targeted to the variable (V) genes (forward primers) and joining (J) genes (reverse primers) of the CDR3 region within IG and TR genes. Samples for which a trackable B-cell receptor sequence was not identified were reflexed for analysis of TRG followed subsequently by TRB. Using high-throughput NGS, the index sequence(s) identified in the diagnostic sample were specifically searched for and quantified, if present, in the remission bone marrow. This assay has an analytic sensitivity of 0.0001% (1 × 10−6), with a sensitivity that is dependent on the cellular input. NGS-based MRD assessment was performed retrospectively for the primary analyses, and this information was therefore not available to clinicians for decision-making. Analysis of a separate validation cohort of prospectively collected clinical NGS MRD assessments was also performed (see section below: “Validation of discordance between RT-PCR for BCR::ABL1 and NGS MRD in prospectively collected clinical samples”), and this information was available in real time to the treating clinician. These latter samples may have been obtained from either the bone marrow or peripheral blood. For all analyses comparing PCR and NGS MRD assessments, samples for both assays were collected at the same timepoint. A flowchart of the study and patient samples in the two cohorts is shown in Figure S1.
2.4 Response and outcome definitions
CMR was defined as the absence of a quantifiable BCR::ABL1 transcript by RT-PCR, as previously described.3 Relapse was defined as recurrence of bone marrow blasts >5% or extramedullary ALL. Relapse-free survival (RFS) was calculated from the time of response until relapse or death from any cause, censored if the patient was alive at last follow-up. Overall survival (OS) was calculated from the time of treatment initiation until death from any cause, censored if the patient was alive at last follow-up. Survival estimates were not censored at the time of alloSCT.
2.5 Statistical methods
Patient characteristics were summarized using median (range) for continuous variables and frequencies (percentages) for categorical variables. To compare two groups with continuous variables, the Wilcoxon rank-sum test was performed. The Kaplan–Meier method was used to estimate the probabilities for RFS and OS and differences between groups were evaluated with the log-rank test. All statistical analyses were performed using GraphPad Prism 9.
3 RESULTS
3.1 Study population characteristics
We identified 44 patients with Ph+ ALL who underwent frontline ALL therapy at our institution, met the study inclusion criteria, and had adequate banked pretreatment samples for NGS MRD analysis. Their baseline characteristics are shown in Table S1. The BCR::ABL1 isoform was p190 in 30 patients (68%) and p210 in 14 patients (32%). The frontline TKI used was imatinib in 1 patient (2%), dasatinib in 24 patients (55%), and ponatinib in 19 patients (43%). Six patients (21%) underwent alloSCT in first remission, all of whom were in the early MRD group. The median follow-up for the entire study population is 84 months.
All baseline samples were calibrated and therefore were trackable for NGS MRD assessment. Among the baseline samples for the 44 patients, 42 were calibrated on BCR, 1 calibrated on TRG, and 1 calibrated on TRB. The median number of dominant sequences per baseline sample was 2 (range, 1–6).
3.2 Correlation of BCR::ABL1 RT-PCR and NGS MRD in the early MRD group
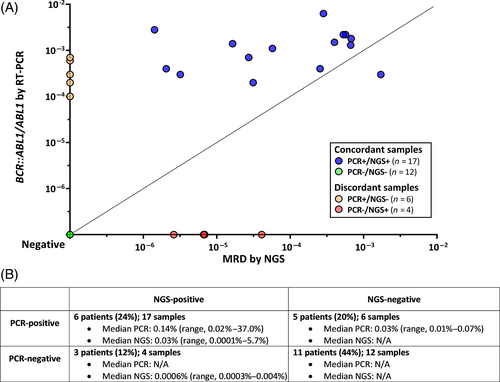
Among the 29 patients who had NGS MRD assessment in the first 6 months of treatment, 46 samples were evaluated for NGS MRD. Seven samples were indeterminate for NGS MRD at a sensitivity of 10−6 (due to insufficient DNA input). In 16 of 17 samples that were PCR+/NGS+ (94%), the PCR value was higher than the NGS MRD value (median level of RT-PCR for BCR::ABL1 0.14% [range, 0.02–37.0%] versus median level of NGS MRD 0.03% [range, 0.0001%–5.7%], respectively; p = .001) (Figure 1A). Among 25 patients with sufficient samples for NGS MRD assessment at 10−6, 8 patients (32%) had discordance between PCR and NGS (5 patients with PCR+/NGS− and 3 patients with PCR−/NGS+) (Figure 1B). Five of 16 patients (31%) who achieved NGS MRD negativity also had detectable BCR::ABL1 by RT-PCR at the same timepoint, four of whom had p190 disease. In the six PCR+/NGS− samples, the median PCR value was 0.03% (range, 0.01%–0.07%). In the four PCR−/NGS+ samples, the median NGS MRD value was 0.0006% (range, 0.0003%–0.004%), in all cases below the level of sensitivity of the PCR assay.
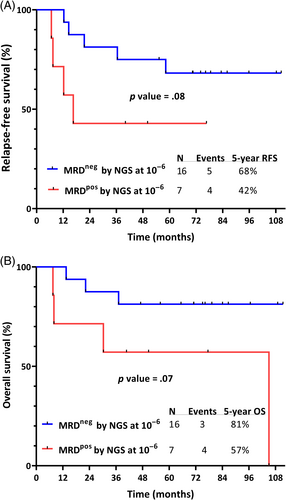
Patients who achieved NGS MRD negativity within the first 6 months of therapy had a trend toward superior RFS compared with those who were NGS MRD positive (5-year RFS 68% and 42%, respectively; p = .08) and superior OS (5-year OS 81% and 57%, respectively; p = .07) (Figure 2A,B). Best PCR response within 6 months of therapy had similar discrimination for RFS and OS (Figure S2); however, numbers were limited to assess the outcomes of patients with discordant MRD assessment by PCR and NGS at these early timepoints.
3.3 Correlation and impact of NGS MRD in the long-term PCR group
One remission sample was evaluated for NGS MRD in each of the 20 patients who were in long-term CMR. The median time from the start of therapy to assessment for NGS MRD was 30 months (range, 26–109 months) and the median follow-up after NGS MRD assessment was 58 months (range, 11–116 months). Eighteen patients (90%) were NGS MRD negative at a sensitivity of 10−6; two patients were indeterminate for NGS MRD at this sensitivity, and no patients were NGS MRD positive. Only 1 of the 20 patients (5%) with long-term CMR relapsed. This patient had received frontline hyper-CVAD plus ponatinib and achieved CMR after the second cycle of treatment. Approximately 1 year after NGS MRD negativity, ponatinib was discontinued due to toxicity and the patient remained off TKI therapy; the patient relapsed 16 months later.
3.4 Correlation and impact of NGS MRD in the long-term PCR+ group
Among the eight patients with long-term PCR positivity, 21 samples were evaluated for NGS MRD, with a median of 2.5 samples per patient (range, 1–4). The median time from the start of therapy to assessment of NGS MRD was 34 months (range, 25–85 months).
Six patients (75%) were NGS MRD negative or indeterminate at a sensitivity of 10−6 at all timepoints assessed (13 samples negative and 4 indeterminate). Five of six patients (83%) with discordant MRD assessment (i.e. PCR+/NGS−) had p190 Ph+ ALL. Among the 13 PCR+/NGS− samples in these 6 patients, the median PCR value was 0.03% (range, 0.01%–17.68%). With a median follow-up of 70 months (range, 11–100 months) since NGS MRD assessment, none of the six patients who were PCR+/NGS− at these later timepoints subsequently relapsed. In these patients, the vast majority (76%–100%) of post-remission PCR values were positive and were generally stable across the treatment course, regardless of any changes in therapy. Notably, two PCR+/NGS− patients underwent therapeutic interventions due to concern for persistent PCR positivity, none of which resulted in CMR. A representative patient is shown in Figure 3A. Due to concern for persistent MRD, this patient received ponatinib plus blinatumomab and then investigational CD19 chimeric antigen receptor T-cells, both without notable change in PCR values. This patient remains in ongoing remission for >60 months, despite persistent low-level PCR positivity (range, 0.01%–0.17%). The other patient received ponatinib plus blinatumomab without significant decline in PCR values (which persisted between 0.01% and 0.02%) and remains in continuous remission for >50 months.
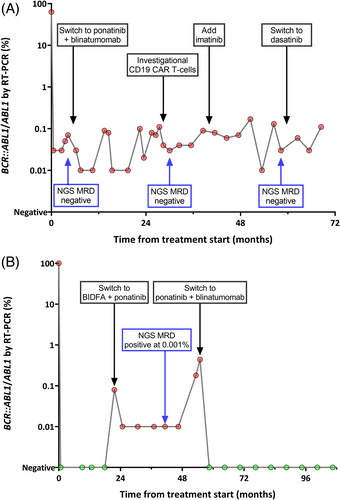
Two patients (25%) were NGS MRD positive at a sensitivity of 10−6, with a median NGS MRD level of 0.001% (range, 0.0004%–0.006%); the median level of MRD by PCR at these timepoints was 0.04% (range, 0.01%–0.07%). In contrast with the PCR+/NGS− group in whom most post-remission PCRs had been positive, the low-level PCR values in these two patients with PCR+/NGS+ MRD were preceded by a period of PCR negativity earlier in the treatment course. Therapeutic interventions following the PCR+/NGS+ timepoint were performed for both patients due to concern for MRD relapse, and both subsequently achieved CMR. A representative patient is shown in Figure 3B. This patient received ponatinib plus blinatumomab due to rising PCR (up to 0.44%), achieved CMR, and remains in continuous CMR for >5 years. The other patient received ponatinib plus blinatumomab for persistent PCR positivity, achieved CMR, but relapsed 8 months later.
3.5 Validation of discordance between RT-PCR for BCR::ABL1 and NGS MRD in prospectively collected clinical samples
To better understand the frequency and prognostic significance of discordant results between RT-PCR and NGS MRD assessment, we reviewed patients with Ph+ ALL at our institution who underwent frontline therapy since June 2021 when we first instituted routine, prospective clinical assessment of NGS-based MRD. Among 74 patients, 65 achieved NGS MRD negativity at a sensitivity of 10−6 at some point over the course of therapy. Among 163 sample timepoints (95 [58%] from the bone marrow and 68 [42%] from the peripheral blood) across these 65 patients, 36 timepoints (22%) were discordant (i.e. PCR+/NGS−), and overall, 11 of 65 patients (17%) who achieved NGS MRD negativity also had detectable BCR::ABL1 transcripts by PCR at the same timepoint. Baseline characteristics of these 11 PCR+/NGS− patients are shown in Table S2. Ten patients (91%) had p190 disease, and 1 patient had p210 disease. The median PCR value at the time of PCR+/NGS− discordance was 0.05% (range, 0.01%–1.23%). Two patients underwent therapeutic intervention due to concern for persistent PCR positivity. One patient received ponatinib plus inotuzumab ozogamicin without notable decline in PCR values (which continued to range 0.06%–1.2%); this patient remains PCR+/NGS−, now on ponatinib monotherapy. The other patient received ponatinib, venetoclax, and blinatumomab but continued to have stable PCR values ranging from 0.07%–0.36%; this patient remains PCR+/NGS− and continues to receive ponatinib plus venetoclax. With a median follow-up of 11 months (range, 1–18 months) since first assessment of PCR+/NGS− discordance, none of the 11 patients have relapsed.
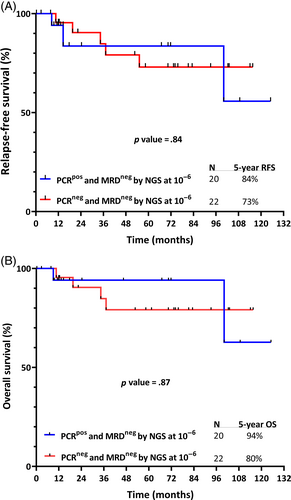
Combining data from patients in both cohorts, we identified 20 patients who were PCR+/NGS− and 22 patients who were PCR−/NGS−. In this combined group, the median follow-up from first documentation of NGS MRD negativity was 61.4 months (range, 1–125 months). RFS (calculated from the first documentation of NGS MRD-negative response) was similar for the PCR+/NGS− and PCR−/NGS− groups, (5-year RFS 84% and 73%, respectively; p = .84); OS was also similar (5-year OS 94% and 80%, respectively; p = .87) (Figure 4). Overall, 3 of the 42 patients (7%) who achieved NGS MRD negativity subsequently relapsed (two in the PCR+/NGS− group and one in the PCR−/NGS− group). These patients all relapsed in the setting of substantial interruptions to TKI therapy (one with multiple, intermittent interruptions due to adverse events and two with prolonged, continuous interruption due to nonadherence).
4 DISCUSSION
Assessment of MRD is prognostically important in Ph+ ALL.3, 6 Persistence of BCR::ABL1 as assessed by RT-PCR is commonly used in clinical practice to identify patients with Ph+ ALL in whom alloSCT in first remission or MRD-directed therapies such as blinatumomab should be considered, particularly in countries where standardized PCR assays for IG/TR are not available.2 However, in this study, we observed that RT-PCR for BCR::ABL1 may be a suboptimal method of MRD assessment in many cases. One-quarter of patients had discordance between PCR and NGS within the first 6 months of frontline therapy, with both PCR−/NGS+ and PCR+/NGS− cases observed. A majority of patients with long-term PCR positivity were NGS MRD negative, despite the NGS MRD assay being 1- to 2-logs more sensitive than the PCR assay. Importantly, these patients did not subsequently relapse, suggesting that NGS MRD provided better discrimination of risk of relapse than did PCR in these cases. Together, these data suggest that NGS MRD may provide more valuable prognostic information than RT-PCR for BCR::ABL1 and that therapeutic decisions in Ph+ ALL may be better informed by also considering NGS MRD status.
In the primary cohort, 5 of 16 patients (31%) who achieved NGS MRD negativity had detectable BCR::ABL1 by RT-PCR at the same time, and in the validation cohort, 11 of 65 patients (17%) who achieved NGS MRD negativity had detectable BCR::ABL1 by RT-PCR. Together, these data suggest that approximately 15–30% of adults with Ph+ ALL who achieve NGS MRD negativity at a sensitivity of 10−6 may still have detectable BCR::ABL1 transcripts by RT-PCR, despite the lower sensitivity of the PCR assay. When limited to patients with BCR::ABL1 PCR positivity >2 years after the start of therapy, the rate of PCR+/NGS− discordance was high (6 of 8; 75%), suggesting that this phenomenon is common in patients with long-term PCR positivity. Other investigators have described patients with this discordance between MRD assessment of BCR::ABL1 and IG/TR as having a “CML-like biology”.5, 8 Using a cell sorting assay, BCR::ABL1 signals can be seen in the nonblast compartment, including in cells of myeloid lineage. This phenotype is commonly observed in patients with p190 Ph+ ALL, and notably, in our validation cohort of 11 patients with PCR+/NGS− MRD, 10 patients (91%) had p190 disease. This entity of Ph+ ALL with CML-like biology therefore is differentiated from CML in lymphoid blast phase, which predominantly carries the p210 BCR::ABL1 isoform.18
We observed that NGS MRD was prognostic for RFS and OS in Ph+ ALL, a finding consistent with several other studies evaluating NGS MRD in Ph-negative ALL across a variety of clinical contexts.9-12, 15 However, given the limited number of patients with adequate banked specimens in our survival analyses, we could not directly compare survival outcomes of patients with discordant MRD assessment by NGS and PCR. Despite this limitation, several observations provide evidence for possibility superiority of NGS MRD in predicting relapse risk in Ph+ ALL. In the primary cohort, we identified six of eight patients with long-term PCR positivity who were PCR+/NGS− at these later timepoints. Despite persistent low-level detectable BCR::ABL1 by RT-PCR, none of these PCR+/NGS− patients had relapsed after a median follow-up of 70 months. A similar finding was observed in our validation cohort, in which none of the 11 PCR+/NGS− patients have relapsed, albeit with a shorter median follow-up. Combining data from both cohorts, we observed that RFS and OS were similar between patients who were PCR+/NGS− and those who were PCR−/NGS−. These observations strongly suggest that achievement of NGS MRD negativity can identify patients with very low risk of relapse, despite persistently detectable BCR::ABL1 by RT-PCR; conversely, since many patients with low-level BCR::ABL1 do not relapse, these findings challenge the notion that achievement of a CMR is universally desirable response endpoint in Ph+ ALL.2 It is also notable that in the two patients who were PCR+/NGS+ at later timepoints, the positive NGS finding appeared to provide evidence that the detectable BCR::ABL1 represented true, clinically significant MRD, as one of these patients experienced hematological relapse and the other experienced a sharp rise in PCR values (i.e. molecular relapse) that responded to ALL-directed therapy. Together, these findings suggest that NGS MRD can further risk stratify patients with positive MRD as assessed by RT-PCR for BCR::ABL1.
Our data have important clinical implications for the treatment of patients with Ph+ ALL. Achievement of CMR is prognostic in Ph+ ALL and has been used as a primary endpoint in several clinical trials.3, 17, 19-22 Persistent MRD by RT-PCR for BCR::ABL1 is also commonly used in clinical practice to identify patients who should be considered for blinatumomab or allogeneic alloSCT.4, 23, 24 Our data suggest that persistent BCR::ABL1 should not be used in isolation as a justification for escalation of therapy, as many of these patients have no detectable disease using a highly sensitive NGS-based MRD assay and have durable remissions and survival. We found that therapeutic interventions for these PCR+/NGS− patients generally do not result in appreciable changes in PCR values, likely because the persistent BCR::ABL1 is present in a nonleukemic clone that is not sensitive to standard ALL-directed therapies. While theoretically one could postulate that these BCR::ABL1-containing nonleukemic cells could predispose to later relapses, it is notable that we did not observe relapses in patients with long-term PCR+/NGS−, despite a relatively long duration of follow-up. This finding is consistent with a report from another group that patients with this CML-like biology had similar outcomes to patients with typical Ph+ ALL and might even have lower rates of relapse.5 Finally, as new and more effective therapies are developed and fewer patients with Ph+ ALL are being referred to alloSCT in first remission, how long patients must receive continuous TKI therapy is an increasingly important clinical question. In many commonly used regimens, BCR::ABL1 TKI therapy is continued for at least 5 years (and in some cases, indefinitely); however, shorter durations of TKI therapy may be possible for some patients with rapid and deep response using highly sensitive MRD assays. The potential for treatment-free remissions in such patients could follow the paradigm established in the treatment of CML, with carefully monitored TKI discontinuation potentially being safe in patients with excellent responses.25
In conclusion, the use of a highly sensitive NGS MRD assay for IG/TR provides important prognostic information in patients with Ph+ ALL and may be better able to guide therapeutic decision-making than does conventional RT-PCR for BCR::ABL1. The use of this NGS assay identified patients with low-level detectable BCR::ABL1 who had very low risk of relapse and did not appear to benefit from therapeutic interventions. Our findings suggest that highly sensitive NGS-based assays could eventually supplant MRD monitoring by RT-PCR for BCR::ABL1 in Ph+ ALL, a question that should be addressed in future prospective studies.
AUTHOR CONTRIBUTIONS
Nicholas J. Short designed the study, collected and analyzed the data, treated patients, and wrote the first draft of the manuscript. Hagop Kantarjian, Farhad Ravandi, Nitin Jain, Fadi G. Haddad, Musa Yilmaz, Ghayas C. Issa, and Partow Kebriaei treated patients. Rashmi Kanagal-Shamanna, Keyur P. Patel, and Sanam Loghavi performed and interpreted standard of care MRD analyses. Walid Macaron and Rebecca Garris collected and analyzed the data and created the tables and figures. Steven M. Kornblau, Sarah Pelletier, Wilmer Flores, and Jairo Matthews collected and distributed banked samples. Elias Jabbour designed the study and treated patients. All authors critically reviewed the manuscript and approved the final version.
FUNDING INFORMATION
Adaptive Biotechnologies Co. performed the NGS MRD assay at no cost to the authors. Supported by an MD Anderson Cancer Center Support Grant (CA016672) and SPORE. N.J.S. is supported by the American Society of Hematology Junior Faculty Scholar Award in Clinical Research.
CONFLICT OF INTEREST STATEMENT
N.J.S has received honoraria from Adaptive Biotechnologies Co. E.J. has received research funding and has received honoraria from Adaptive Biotechnologies Co. The other authors report no relevant conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data available on request from the authors.




