Attenuation of the Fish Pathogen Francisella sp. by Mutation of the iglC* Gene
Abstract
Fish francisellosis is an emergent disease caused by gram-negative facultative intracellular bacteria of the genus Francisella. Different strains of the bacterium have caused high mortalities in warmwater and coldwater fish species. Francisella sp. isolates from fish have been found to share more than 97% identity to the human pathogen Francisella tularensis upon 16S ribosomal RNA sequence comparison. Homologue genes of the F. tularensis intracellular growth locus (iglA*, iglB*, iglC*, and iglD*) were identified from LADL 07-285A, a clinical isolate obtained from diseased Nile tilapia Oreochromis niloticus. The iglABCD operon DNA sequence comparison revealed that Francisella LADL 07-285A had 94% identity with F. philomiragia subsp. philomiragia and 83% identity with F. tularensis subsp. novicida U112. The functions of the conserved proteins corresponding to the genes are elusive but appear to be essential for the ability of Francisella sp. to survive within macrophages and cause disease. An insertion mutation was made in the iglC* gene of LADL 07-285A by allelic exchange, and the iglC* mutant was found to be attenuated after intraperitoneal and immersion challenges in Nile tilapia. Laboratory challenge methods for inducing francisellosis in Nile tilapia were evaluated by intraperitoneal injection and immersion with serial dilutions of Francisella LADL 07-285A. The dose lethal to 50% of test fish at 40 d postchallenge was 10−5.3 (about 1.2 × 103 colony-forming units/fish) by intraperitoneal injection and was 10−1 (2.3 × 107 colony-forming units/mL of tank water) by immersion.
Tilapias are among of the most important cultured fish species in the world. Worldwide aquaculture production of tilapias, mainly Nile tilapia Oreochromis niloticus, has been increasing in exponential proportions during the last decade to greater than 2.5 × 106 metric tonnes in 2005. The main producing countries are China, Ecuador, Egypt, Israel, Indonesia, Singapore, Philippines, and Thailand, but Mexico, Costa Rica, Honduras, and other Latin American countries have more than doubled their production in the past 5 years. The USA is the country that imports the highest amount of tilapia, receiving more than 80% of worldwide tilapia exports (Josupeit 2008). As the tilapia aquaculture industry expands, tilapia farms are often challenged with disease outbreaks, which in several cases have caused severe economic losses due to high mortality events, decreased weight gain, and antibiotic and treatment expenses.
Francisella sp. is an emergent bacterial pathogen that causes acute to chronic symptoms in cultured and wild fish belonging to warmwater and coldwater species. During the past 5 years, Francisella sp. has been implicated as the cause of mortalities in tilapia and other important warmwater and coldwater species cultured in the USA, Taiwan, Costa Rica, Latin America, Hawaii, Norway, Chile, and Japan (Kamaishi et al. 2005; Hsieh et al. 2006; Ostland et al. 2006; Birkbeck et al. 2007; Mauel et al. 2007; Mikalsen et al. 2007; Ottem et al. 2007; Soto et al., in press). Infected fish present with nonspecific clinical signs, such as erratic swimming, anorexia, anemia, exophthalmia, and high mortality. Upon gross and microscopic examination, several internal organs (mainly spleen and kidney) are enlarged and contain widespread multifocal white nodules. Histological examination reveals the presence of multifocal granulomatous lesions, and numerous small, pleomorphic cocco-bacilli are present (Soto et al., in press).
Although species of Francisella have been identified as emerging pathogens of fish, many are poorly characterized due to the fastidious nature of the bacteria and difficulties in culturing these organisms from fish tissues; for this reason, many have not been properly classified. In the majority of the cases, polymerase chain reaction (PCR) and sequence comparison of the 16S ribosomal RNA have made it possible to place the organism at 97% similarity to the human pathogen F. tularensis, 98% similarity to F. philomiragia, and 99% to other strains isolated from fish species (Kamaishi et al. 2005; Hsieh et al. 2006; Ostland et al. 2006; Mauel et al. 2007; Mikalsen et al. 2007; Ottem et al. 2007; Soto et al., in press). Francisella philomiragia subsp. noatunensis and F. piscicida were recovered from moribund farmed Atlantic cod Gadus morhua in Norway (Mikalsen et al. 2007; Ottem et al. 2007), which displayed chronic granulomatous disease. Strains from Atlantic cod in Norway have been characterized by phenotypic and molecular taxonomic methods as closely related members of F. philomiragia subsp. philomiragia (Mikalsen et al. 2007; Ottem et al. 2007).
Currently, it is not known if the isolate recovered from diseased Nile tilapia in Costa Rica (Soto et al., in press) and the Atlantic cod isolates represent two different bacteria species or two subspecies of the same bacterium. The strain used in this research project, LADL 07-285A, was isolated at the Louisiana Aquatic Diagnostic Laboratory, Louisiana State University School of Veterinary Medicine, Baton Rouge, from Nile tilapia sampled in Costa Rica; this isolate was confirmed by molecular analysis as Francisella sp. It exhibited 99% identity with other fish pathogenic species of Francisella (Soto et al., in press).
The most important species in the genus is F. tularensis (Dennis et al. 2001; Sjostedt 2007). Besides being an important animal pathogen, F. tularensis is a zoonotic agent that has received considerable study as a potential bioterrorism agent. Francisella tularensis has a high infectivity rate and multiple infectious routes (Keim et al. 2007; Nano and Schmerk 2007). The genetic basis of F. tularensis virulence is still poorly understood, although several virulence determinants have been identified (Golovliov et al. 2003; Nano et al. 2004; Barker and Klose 2007). Baron and Nano (1998), Golovliov et al. (2003), and de Bruin et al. (2007) have described the intracellular localization, survival, replication, and escape of F. tularensis subspecies in adherent mouse peritoneal cells, a mouse macrophage-like cell line (J774A.1), and a human macrophage cell line (THP-1). Some of the most interesting genes identified in F. tularensis are the genes of the intracellular growth locus (IGL; iglA*, iglB*, iglC*, and iglD*) present as part of a 30-kilobase-pair pathogenicity island described by Nano et al. (2004) and Barker and Klose (2007). The functions of the conserved proteins corresponding to the genes are elusive. Overall, IGL proteins appear to be essential for the ability of F. tularensis to survive inside the macrophages and cause disease (Golovliov et al. 1997; Lai et al. 2004; Lauriano et al. 2004; Nano et al. 2004; Santic et al. 2005; Brotcke et al. 2006; de Bruin et al. 2007). Recent data have shown that iglA* and iglB* are part of a novel type-6 secretion system encoded by the Francisella pathogenicity island (Nano and Schmerk 2007; Ludu et al. 2008a). Mutations of these four genes in F. tularensis have shown decreased pathogenicity of the bacterium, both in vivo and in vitro, in mammalian and insect tissues and cell lines (Lauriano et al. 2003; Nano et al. 2004; de Bruin et al. 2007; Vonkavaara et al. 2008).
This research examined the presence of homologues to the igl* virulence genes in fish pathogenic Francisella sp. We developed a useful method for allelic exchange using PCR products to mutate Francisella sp. and to create an attenuated iglC* mutant upon intraperitoneal (IP) and immersion challenges in Nile tilapia. Finally, we conducted IP and immersion infectivity trials to induce francisellosis in Nile tilapia, and here we report the dose required to cause mortality in 50% of the fish (LD50) after challenge with this important emergent fish pathogen.
Methods
Bacterial Strains and Growth Conditions
Strains, plasmids, and primers used in this study are listed in Table 1. Francisella LADL 07-285A was isolated from cultured Nile tilapia by our laboratory and described in previous work (Soto et al., in press). Francisella LADL 07-285A was grown for 48 h at 28°C in cystine heart agar supplemented with bovine hemoglobin solution (CHAH; Becton, Dickinson, and Company, BD, Sparks, Maryland). A liquid culture medium consisted of a modified Mueller-Hinton II (MMH) cation-adjusted broth supplemented with 2% IsoVitaleX (Becton, Dickinson, and Company, BBL) and 0.1% glucose (Soto et al., in press). Broth cultures were grown overnight at 25°C in a shaker at 175 revolutions/m, and bacteria were frozen at −80°C in the broth media containing 20% glycerol for later use. Polymixin B (100 units/mL) and ampicillin (50 μg/mL) were added when needed to make the primary isolation media selective, which aided in recovery of the bacteria from fish tissues; kanamycin (15 μg/mL) was used for recovery of transformed bacteria following electroporation. We grew Escherichia coli XL1 Blue MRF by using Luria-Bertani broth or agar for 16–24 h at 37°C and supplemented with kanamycin (50 μg/mL), when needed, to recover the plasmid containing bacteria after electroporation.
Identification of iglABCD operon homologue
The complete genome sequences of F. philomiragia subsp. philomiragia ATCC (American Type Culture Collection) 25017 (GenBank accession number CP000937), F. tularensis subsp. novicida U112 (GenBank CP000439), and partial genome sequences of F. piscicida strain GM2212 (GenBank EU492905), available from the National Center for Biotechnology Information (NCBI), were used to compare the iglABCD regions. Previously published F. tularensis primers to these genes were also compared and were used as templates to design primers for PCR amplification of homologous regions from the chromosomal DNA of Francisella LADL 07-285A. These PCR amplicons for sequencing were purified with the QiaQuick Minelute PCR Cleanup Kit (Qiagen, Valencia, California) as directed by the manufacturer and were sequenced on an Applied Biosystems 3130 Genetic Analyzer via the PCR primers described in Table 1.
The sequences from the four Francisella LADL 07-285A iglABCD genes and the corresponding amino acid sequences were compared with those stored in the NCBI database via the BLASTN and BLASTP programs using default settings.
Electroporation
Electrocompetent Escherichia coli and Francisella LADL 07-285A were prepared following Maier et al. (2004) with some modifications. Briefly, E. coli was aerobically grown until mid-logarithmic stage (optical density at 600 nm [OD600] = 0.7), and the cells were prepared by washing two times in water followed by one wash in 10% glycerol. The electrocompetent E. coli were electroporated using a BioRad Gene Pulser Controller in a 2-mm electroporation cuvette (BTX Harvard, Holliston, Massachusetts). The pulser was set at a voltage of 2.5 kV, a capacitance of 25 microfarads (μF), and a resistance of 200 Ω. Immediately after electroporation, cells were suspended in 1 mL of Luria-Bertani broth and incubated with shaking for 1 h at 37°C. After the 1-h incubation period, E. coli was plated on Luria-Bertani agar with kanamycin (50 μg/mL).
We grew Francisella LADL 07-285A aerobically until the late-logarithmic stage (OD600 = 0.6); the cells were prepared by using 0.5-M sucrose. The electrocompetent Francisella sp. was electroporated using the BioRad Gene Pulser in a 2-mm cuvette. The pulser was set at a voltage of 2.5 kV, a capacitance of 25 μF, and a resistance of 600 Ω. Immediately after electroporation, cells were suspended in 1 mL of MMH broth and incubated with shaking for 4 h at 28°C. After the 4-h incubation period, Francisella sp. was plated on CHAH with kanamycin (15 μg/mL).
Mutant and plasmid construction
A fragment of approximately 850 base pairs (bp) corresponding to a portion of the iglB* and iglC* genes from Francisella LADL 07-285A was PCR-amplified via primers F-40 and F-41 (Table 1), which contain XhoI and SpeI sites, respectively. All enzymes used during the study were supplied by New England Biolabs and were used under the conditions recommended by the manufacturer. The PCR product was cleaved with these two endonucleases and was ligated into the high copy number plasmid, pBluescript SK (pBS), resulting in the plasmid pBS-iglC*. The plasmid was electroporated into E. coli, amplified, and then purified from the bacterium via the QIAprep Spin Miniprep Kit (Qiagen) following the manufacturer's protocol.
Plasmid pEN1, constructed and donated by Ludu et al. (2008b), contains a Tn903 kanamycin cassette (Km-P) linked to a Francisella novicida promoter derived from the region upstream of gene FTN1451* (Gallagher et al. 2007). Purified pEN1 plasmid was digested with PstI to release the Km-P cassette.
For the construction of pBS-ΔiglC*, plasmid pBS-iglC* was digested with PstI endonuclease, which cuts once in the iglC*gene. The 1,100-bp KmP cassette was ligated into the unique PstI site in pBS-iglC*, resulting in pBS-ΔiglC*. The resulting insertion was verified by sequencing.
Nile Tilapia LD50 Virulence Assays
Preparation of bacterial stock culture and enumeration
We cultivated Francisella LADL 07-285A in MMH in a shaking incubator at 175 revolutions/min overnight at 25°C. The bacteria were then pelleted, and the concentrations were adjusted to about 2.3 × 109 colony-forming units (CFU) per milliliter in phosphate-buffered saline. Tenfold dilutions of this stock were then made in sterile saline. Actual bacterial numbers delivered by injection and by immersion were determined by colony counts on CHAH plates. Enumeration of the bacteria was done by placing 50-μL drops of each 10-fold dilution on CHAH and counting the resulting colonies after 72 h of incubation at 25°C. The dilution, which produced countable colonies (10–50 colonies/drop), was then used to calculate the concentration (CFU/mL) in the stock suspension.
Fish and systems
Naïve Nile tilapia (average length, about 9.0 cm; average weight, about 18.9 g) were obtained from a source believed to be free of Francisella infection. A subsample of the population was confirmed as negative for Francisella sp. bacteria by culture on CHAH and PCR before use in the study. Groups of 10 fish were placed in 90-L tanks with filtered, recirculating water flow (1 tank/treatment). Water temperature was maintained in the range of 23–25°C throughout the study, and fish were fed daily with a commercial 4.7-mm pelleted fish feed (Cargill, Franklinton, Louisiana) at 5% of body weight. The fish were allowed to acclimate for at least 2 weeks before challenge. At challenge, all fish were anesthetized with tricaine methanesulfonate (MS-222; 100 mg/L of water) prior to handling.
Intraperitoneal injection
From the initial bacterial concentration in the stock suspension (2.3 × 109 CFU/mL in phosphate-buffered saline), 10 serial dilutions in phosphate-buffered saline were prepared. Each fish received an IP injection of 0.1 mL of the bacterial suspension. Fish in the control tank were injected with 0.1 mL of sterile phosphate-buffered saline. Mortality was recorded daily after IP injection, and the LD50 was calculated for the wild-type strain of Francisella LADL 07-285A.
Immersion challenge
Immersion challenge was carried out in eight different dilutions of the bacterial suspension: 10 L of static water containing 2.3 × 108, 2.3 × 107, 2.3 × 106, 2.3 × 105, 2.3 × 104, 2.3 × 103, 2.3 × 102, 2.3 × 101 CFU/mL of tank water. Each immersion-challenged fish received its respective treatment for 3 h. After 3 h, fish were moved to a clean, 90-L tank system with biofiltered recirculating water. Control fish were treated with sterile phosphate-buffered saline in a similar manner.
Analysis of dead and surviving fish after challenge
Dead and surviving fish were subjected to a complete clinical and bacteriological examination. A histological evaluation was performed on splenic, hepatic, and renal tissue of moribund, freshly dead, and surviving fish. Severity of the disease in each treatment was determined by counting the number of granulomas in histological sections present per single 10× microscopic field from the spleen, head kidney, and liver of each fish, and the means were reported as severe (>20), moderate (7–20), or mild (<7). Molecular analysis by PCR was performed (protocol of Soto et al., in press) on bacterial cultures recovered from moribund and dead fish and on DNA extracted from spleen tissue of fish surviving the challenge. The LD50 at days 20 and 40 after both the IP and the immersion challenges was calculated by the method of Reed-Muench (Anderson 1984).
In Vivo Challenge with Francisella Wild-Type and ΔiglC* Strains
The wild-type and ΔiglC* strains were tested for virulence by both IP and immersion challenge. Francisella LADL 07-285A wild-type and ΔiglC* isogenic strains were grown on CHAH plates at 25°C for 72 h. Cells were harvested, suspended in 1 L of MMH broth, and incubated in a shaking incubator overnight at 24°C to obtain a final OD600 of 0.75. Enumeration of bacteria in IP and immersion challenges was accomplished by the same methods outlined for the LD50 study.
Fish and systems
The fish were obtained from the same source, were in the same size range, and were fed the same way as previously described for LD50 study. The challenge trials were done in 20-L, flow-through tanks but with chlorination traps in the drain system for biosafety. Fish were maintained at 10 fish/tank; four tanks were used per treatment, one tank serving as a noninfected control. Before challenge, all fish were anesthetized with MS-222 (100 mg/L). The IP fish received a 0.1-mL injection of one of two bacterial suspensions: about 3 × 108 or 1.5 × 108 CFU/fish. The immersion-challenged fish were immersed for 3 h in tanks containing 8 L of static water with approximately 3.7 × 107 or 1.8 × 107 CFU/mL. After 3 h, the volume of the tanks was adjusted to a maximum of 20 L with clean, dechlorinated, aerated municipal water. Control fish were treated in a similar fashion but received sterile phosphate-buffered saline in place of the bacterial suspension.
After each challenge exposure, mortality was recorded every 12 h for 30 d. Dead fish and survivors from each challenge were subjected to a complete clinical and bacteriological evaluation. Polymerase chain reaction was performed on DNA from bacterial cultures recovered from moribund and dead fish to confirm the presence of the wild-type or ΔiglC* strain.
Statistical Analysis
Data (both original and inverse-sine transformed) obtained from IP and immersion challenges with the Francisella LADL 07-285A wild-type and ΔiglC* strains were compared in an analysis of variance with a factorial arrangement of treatments by use of the Statistical Analysis System version 9.1.3. Where significance was found, post hoc pairwise comparisons were conducted with t-tests of least-squares means. Differences were considered significant at P-values of 0.05 or less.
Results
Identification of the iglABCD Operon
The deduced amino acid products of the Francisella LADL 07-285A iglA* gene had 95, 92, and 88% similarity to the IGL-A protein of F. philomiragia subsp. philomiragia, F. piscicida, and F. tularensis subspecies, respectively. The amino acid sequences of the Francisella LADL 07-285A proteins IGL-B, IGL-C, and IGL-D showed identities of 97, 95, and 92% (IGL-B); 93, 90, and 89% (IGL-C); and 94, 92, and 80% (IGL-D) to the IGL proteins found in F. philomiragia, F. piscicida, and F. tularensis, respectively. The guanine plus cytosine content found in the iglABCD operon from LADL 07-285A (GenBank FJ386388) was 31%. Overall DNA comparison of the iglABCD operon showed that the fish pathogen Francisella LADL 07-285A shared 94% identity with F. philomiragia subsp. philomiragia and 83% identity with F. tularensis subsp. novicida U112. The iglABCD operons were in the same orientation and arrangement for the three members of Francisella.
Generation of a Francisella LADL 07-285A iglC* Mutant
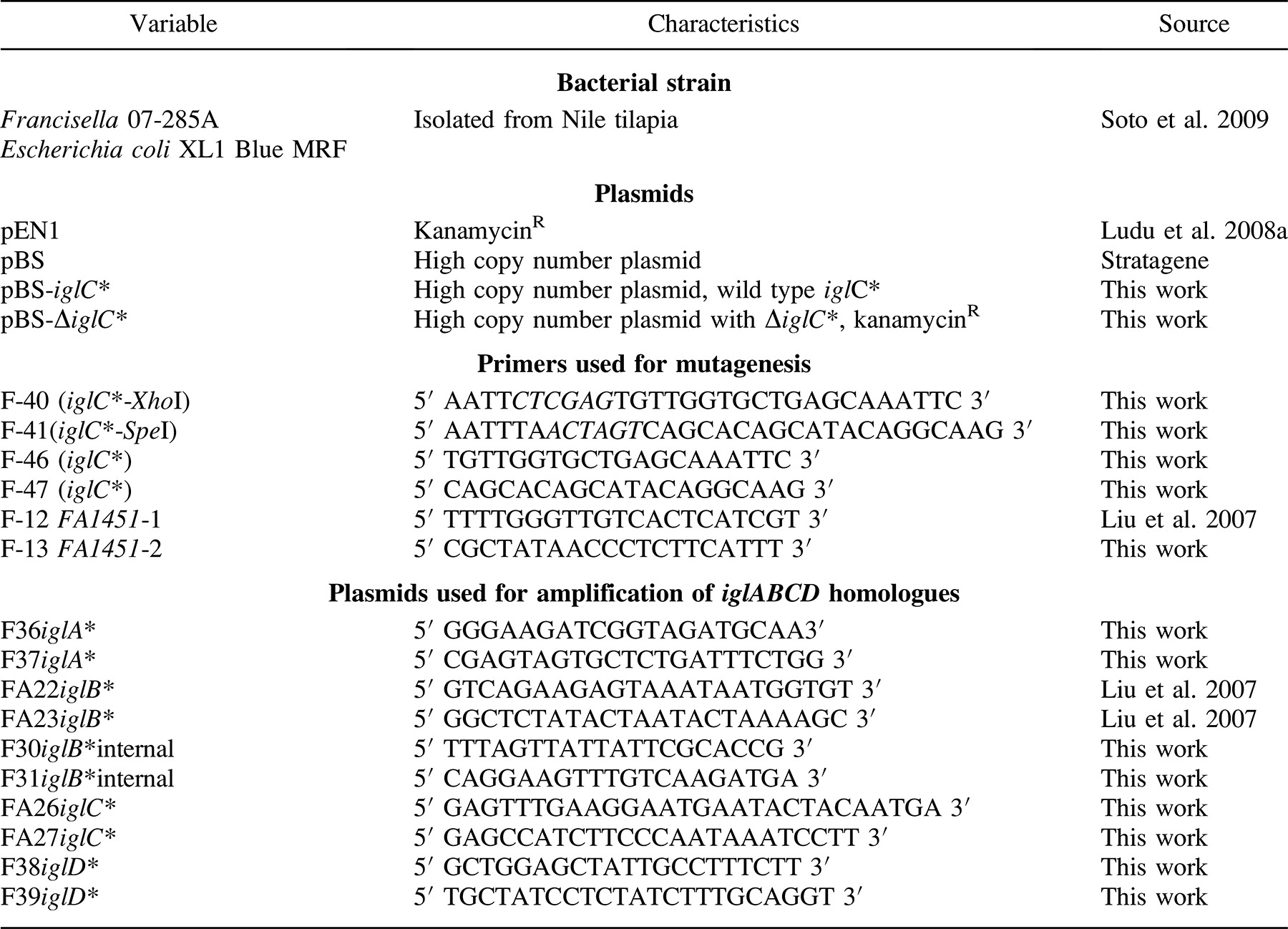
Francisella tularensis IGL-C protein expression has been shown to be upregulated and important for intramacrophage survival and growth in F. tularensis subspecies (Nano et al. 2004; de Bruin et al. 2007). An insertion mutation made in the iglC* gene of LADL 07-285A by allelic exchange using Km-P was found to have approximately 400 bp of flanking sequences on either side of the insertion site. Insertion of Km-P was confirmed by PCR using two different sets of primers and DNA sequencing. Primer sets F-46–F-47 and F-31–F-38 were used to verify the insertion and position of the 1,100-bp Km-P cassette in iglC* (Figure 1). Primers used for amplification of the FA1451* promoter region were also used to sequence the inside region of the insertion and to verify the presence of the promoter in the mutant.
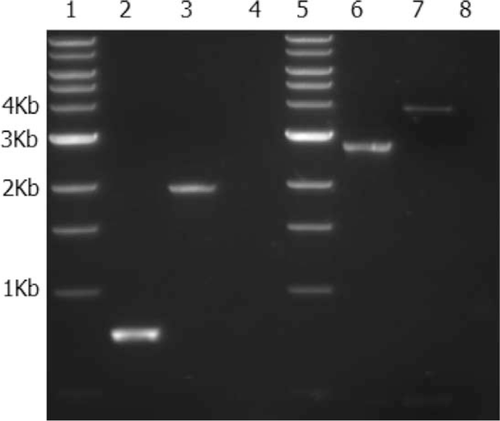
Polymerase chain reaction (PCR) amplification of the iglC* gene from wild-type and isogenic mutant (ΔiglC*) strains of Francisella LADL 07-285A. Lanes 1 and 5 show a 1-kilobase-pair ladder. Lanes 2 and 6 show PCR amplification of the iglC* gene from the wild-type strain; lanes 3 and 7 show PCR amplification of the gene from the ΔiglC* strain. Primer sets were F-46–F-47 for lanes 2 and 3 and F-31–F-38 for lanes 6 and 7. Lanes 4 and 8 are water.
The resulting Francisella LADL 07-285A ΔiglC* strain had no obvious morphological differences from the wild-type strain, and growth characteristics were identical to those of the parental strain in broth and on agar media.
Nile Tilapia LD50 Virulence Assays
Based on the cumulative mortalities, the observed LD50 for the IP-challenged Nile tilapia infected with Francisella LADL 07-285A was 10−5.1 (about 1.8 × 104 CFU/fish) on day 20 and 10−5.3 (about 1.2 × 104 CFU/fish) on day 40 (Figure 2). On the other hand, the observed LD50 for the immersion-challenged Nile tilapia was 10−0.52 (about 6.9 × 107 CFU/mL) at day 20 and 10−1 (about 2.3 × 107 CFU/mL) at day 40 (Figure 3). The least amount of bacteria required to cause mortality in the IP-challenged Nile tilapia was 23 CFU/mL of phosphate-buffered saline, whereas for the immersion-challenged fish, 2.30 × 102 CFU/mL of tank water was necessary to cause mortality (Table 2).

Mortality of Nile tilapia 1–40 d after challenge with Francisella LADL 07-285A via intraperitoneal injection (colony-forming units [CFU]/mL of phosphate-buffered saline; 10 fish were infected per treatment).
Mortality of Nile tilapia 1–40 d after challenge with Francisella LADL 07-285A via immersion (colony-forming units [CFU]/mL of tank water; 10 fish were infected per treatment).
The dead and moribund fish presented the same clinical signs and histopathological lesions as described in Soto et al. (in press). No obvious external clinical signs were observed in the fish. Internally, the most significant gross pathological change observed was the presence of widespread multifocal white nodules dispersed in the anterior kidney, posterior kidney, and spleen, with a marked splenomegaly and renomegaly. Histopathologically, granulomatous inflammation was present in the spleen and kidneys, where large numbers of macrophages contained small pleomorphic cocco-bacilli. We isolated Francisella LADL 07-285A from the spleen and kidney of dead and moribund fish from both treatments (Table 2).
In Vivo Challenge with Francisella LADL 07-285A Wild Type and ΔiglC*
To examine the role of the iglC* gene on virulence in a fish model of infection, we measured survival rates of Nile tilapia infected with Francisella LADL 07-285A wild type and ΔiglC* by two different routes of inoculation. At 48 h after IP injection of 0.1 mL of bacterial suspension (at approximately either 3 × 108 or 1.5 × 108 CFU/fish) all infected Nile tilapia challenged with the wild type had died. Only one fish infected with the ΔiglC* had died by 30 d postchallenge; this fish was not examined because it was in an advanced stage of decomposition. The difference in dosages did not show significance, whereas the percent mortality between wild-type- and mutant-injected fish was significantly different (P < 0.0001; Figure 4).

Percent mortality of Nile tilapia receiving an immersion challenge (IC) or intraperitoneal (IP) injection with the Francisella LADL 07-285A wild-type strain or isogenic mutant strain (ΔiglC*). Bacterial treatments differed by challenge method, bacterial type, and number of colony-forming units (CFU; all approximate: treatment A = IP injection with wild type at 3 × 108 CFU/fish; B = IP injection with ΔiglC* at 3 × 108 CFU/fish; C = IP injection with wild type at 1.5 × 108 CFU/fish; D = IP injection with ΔiglC* at 1.5 × 108 CFU/fish; E = IC with wild type at 3.7 × 107 CFU/mL; F = IC with ΔiglC* at 3.7 × 107 CFU/mL; G = IC with wild type at 1.8 × 107 CFU/mL; H = IC with ΔiglC* at 1.8 × 107 CFU/mL). Asterisks denote significant differences between treatment pairs with differing bacterial types (A versus B; C versus D; E versus F; G versus H; P < 0.0001 for all comparisons).
Fish immersed in tank water containing wild-type bacteria at about 3.7 × 107 CFU/mL had a survival rate of 43.3%, and survival was 56.6% for fish immersed in 1.8 × 107 CFU/mL. The dosages did not result in significantly different mortality. On the other hand, the fish challenged with the mutant strain had a 100% survival rate when challenged with about 3.7 × 107 and 1.8 × 107 CFU/mL. Percent mortality was significantly different between groups challenged with the wild-type and mutant strains (P < 0.0001; Figure 4).
The histopathological analysis of the fish challenged with the wild type showed the same lesions as previously described for the LD50 challenge: increased melanomacrophage centers, widespread granulomas, and granulomatous inflammation in the spleen and head kidney. Upon gross and histopathological analysis, the fish challenged with the ΔiglC* by immersion did not show any granulomatous lesions or increased number of melanomacrophages in the analyzed tissues. The fish challenged with the ΔiglC* strain by IP injection had no granulomas but had higher numbers of melanomacrophages in the head kidney and spleen at 30 d postchallenge than did the control group injected with phosphate-buffered saline. The control fish immersed with phosphate-buffered saline did not display any lesions in the tissues and organs.
Discussion
Members of the genus Francisella are fastidious facultative bacteria that have been found to infect a great variety of animals, including humans, but very little is known regarding the virulence mechanisms and virulence factors of this genus (Barker and Klose 2007; Keim et al. 2007). Subspecies of F. tularensis have been found to exist within macrophages in different vertebrate hosts, arthropods, and in amoebae (Abd et al. 2003; Keim and Wagner 2007; Vonkavaara et al. 2008). Several genes provide the pathogen with properties for survival in the extracellular compartment and also for survival and multiplication inside of potent phagocytes like neutrophils and macrophages (Baron and Nano 1998; Allen 2003; Nano et al. 2004). During previous histopathological analysis of infected tissue from Nile tilapia, we observed the presence of large numbers of bacteria inside the macrophages, implying that the organism was a facultative intracellular pathogen.
The first objective of this study was to investigate the presence of homologous virulence factors in the emergent fish pathogen Francisella sp. and the other species of Francisella. By comparing different Francisella species' genomes, including F. tularensis and F. philomiragia genomes available at the NCBI website, we constructed primers to amplify the iglABCD operon from the Nile tilapia isolate.
The iglABCD operon was described by Nano et al. (2004) as part of the F. tularensis pathogenicity island. The ability to survive inside macrophages and the presence of a type-6 secretion system are two of the most important virulence factors described for the pathogenicity island of this important human pathogen.
In this study, we target a homologue of the highly expressed intracellular protein, IGL-C, in F. tularensis. The insertional mutagenesis protocol followed in this study allowed us to select for a double recombination in the Francisella LADL 07-285A iglC* gene. Kanamycin was used as the selective antibiotic resistance marker because of the natural kanamycin susceptibility of the Francisella sp. strain used in this study (data not shown). As in F. tularensis subspecies, the iglC* mutation significantly attenuates the pathogen upon in vivo challenges and increases the survival rates of the mutant-infected fish compared with the wild-type-infected fish after both IP and immersion challenges. We suspect that the presence of melanomacrophages in the spleen and head kidney of IP-injected fish challenged with the mutant strain is due to a normal immune reaction of fish targeting the injected bacterium and is not due to intracellular survival of the bacterium in macrophages, as was observed in fish infected with the wild-type strain. Current work is being done in our laboratory to determine the pathogenesis of the Nile tilapia isolate after experimental infection, including the fate of cells after phagocytosis by the macrophage. Also, we would like to assess the survival of the bacterium in Nile tilapia macrophages after in vitro cell challenges.
We also compared two different administration routes (IP and immersion) for challenging Nile tilapia with Francisella sp. and reported the LD50s observed at 20 and 40 d postchallenge. The IP challenge was chosen because it was an easy and quick method to accurately administer suspended bacteria, but several problems developed when administering the bacteria by this method, including the lack of exposure of the bacteria to innate immune protection present in the skin, gills, and other mucosa. As was expected, an acute onset of the disease was observed, and high mortalities and few clinical signs occurred in the fish receiving the higher dosage. It was surprising that a low dose of bacteria (about 23 CFU injected into the peritoneum of the fingerlings) was able to cause mortalities. Even more surprising was that even a very low number of bacteria (about 1 CFU/fish) produced a large number of severe lesions (granulomas) in important hematopoietic and osmoregulatory organs like the spleen and the anterior kidney. Survivors of this treatment were observed with significant lesions in spleen, head kidney, and liver; such lesions not only impair the fish's ability to osmoregulate but also cause immunosuppression by direct damage of hematopoietic organs, making the fish more susceptible to other important and common diseases found in culture facilities (e.g., streptococcosis and columnaris disease).
The immersion challenge route was chosen because it more closely resembles a natural infection. The necessity for the bacteria to come into close contact with the innate immune system present in skin, gills, gastrointestinal mucosa, and other organs more closely resembles the way the disease progresses in nature. As expected, the amount of bacteria needed to cause mortality was higher than in the IP treatment, and the onset of the disease was more subacute to chronic, presenting anorexia, change in coloration, and pale gills. A dose of 2.3 × 102 CFU/mL was needed to cause mortality in the immersed fish, but when analyzing histopathological lesions of the survivors, it was evident that even a dose of 23 CFU/mL was able to cause significant lesions in the spleen and head kidney (Figure 5). The experiment was terminated 40 d after exposure to the bacterium, but we suspect that the survivors of this trial could have become carriers of the pathogen, as seen in natural infections.
Histological photomicrographs of two Nile tilapia spleens (stained with hematoxylin and eosin): noninfected spleen (left panel), showing normal splenic parenchyma and stroma; and severely infected spleen (right panel) at 40 d after immersion challenge with Francisella LADL 07-285A (23 colony-forming units/mL), showing widespread multifocal granulomatous lesions with mixed inflammatory infiltrates.
This study provides the first identification of homologous genes of the F. tularensis pathogenicity island in the fish pathogenic Francisella sp. The data gathered in this study provide useful information of potential target genes for future virulence, pathogenesis, and immunological studies of this emergent worldwide pathogen. It also provides an easy and reliable method for mutagenesis of this fastidious pathogen. We also show important data regarding the virulence capacity of the bacterium in Nile tilapia and provide examples of two challenge methods for experimental infection of Nile tilapia. Data from challenge trials indicate that mutation of iglC* results in a less-virulent pathogen, presumably due to decreased intramacrophage survival. This will be assessed in future in vitro studies.
Acknowledgments
We gratefully thank Jagit Ludu and Francis Nano at the Department of Biochemistry and Microbiology, University of Victoria, British Columbia, for sharing some of the plasmids used in this study. We also thank Judy Wiles, Judith Beekman, Matt Rogge, Wes Baumgartner, and Ron Thune of the Pathobiological Sciences Department, Louisiana State University School of Veterinary Medicine, for their skillful technical advice and assistance.