Renal Chloride Channel, CLCN5, Mutations in Dent's Disease
Abstract
Dent's disease is an X-linked renal tubular disorder characterized by low-molecular-weight proteinuria, hypercalciuria, nephrocalcinosis, nephrolithiasis, and renal failure. Patients with Dent's disease may also suffer from rickets and other features of the renal Fanconi Syndrome. Patients may have mutations in the X-linked renal chloride channel gene, CLCN5, which encodes a 746-amino-acid protein with 12–13 transmembrane domains. We have investigated the 11 coding exons of CLCN5 for mutations in eight unrelated patients with Dent's disease. Leukocyte DNA was used for the polymerase chain reaction amplification of CLCN5 and the products analyzed for single-stranded conformational polymorphisms (SSCPs). Abnormal SSCPs were sequenced and revealed eight mutations. These consisted of three nonsense mutations (Arg34Stop, Arg648Stop, Arg704Stop), four deletions involving codons 40, 86, 157, and 241, and one acceptor splice consensus sequence mutation tgcag → tgaag. The mutations were confirmed either by restriction endonuclease or sequence-specific oligonucleotide hybridization analysis. In addition, an analysis of 110 alleles from 74 unrelated normal individuals demonstrated that the DNA sequence changes were not common polymorphisms. All of the mutations predict truncated chloride channels that are likely to result in a functional loss. Thus, our findings expand the spectrum of CLCN5 mutations associated with Dent's disease and the results will help to elucidate further the functional domains of this novel chloride channel.
INTRODUCTION
DENT'S DISEASE, which is a disorder of mineral homeostasis that is due to renal tubular dysfunction, is characterized by low-molecular-weight proteinuria, hypercalciuria, nephrocalcinosis, nephrolithiasis, and eventual renal failure.1, 2 Dent's disease may also be associated with aminoaciduria, phosphaturia, glycosuria, kaliuresis, uricosuria, and impaired urinary acidification and is often complicated by rickets or osteomalacia. Thus, Dent's disease is a form of the Fanconi Syndrome.2, 3 However, the common occurrences of hypercalciuria, nephrocalcinosis, and kidney stones in Dent's disease and the unusual or rare association of these with the Fanconi Syndrome, are important differences between these two disorders. Dent's disease has features in common with X-linked recessive nephrolithiasis,4, 5 X-linked recessive hypophosphataemic rickets,6 and the idiopathic low-molecular-weight proteinuria of Japanese children.7 All of these four disorders have been shown to be due to inactivating mutations of the voltage-gated chloride channel gene CLCN5,8-12 and on the basis of the phenotypic similarities and the common genetic etiology these are collectively referred to as Dent's disease.9 The human CLCN5 gene, which is located on chromosome Xp11.22, has a 2238-bp coding sequence that consists of 11 exons which span 25–30 kb of genomic DNA and encode a 746 amino acid protein (Fig. 1).13, 14 CLCN5 belongs to the family of voltage-gated chloride channel genes (CLCN1-CLCN7, and CLCKa and CLCKb) that have ∼12 transmembrane domains.15 These chloride channels have an important role in the control of membrane excitability, transepithelial transport, and possibly cell volume.15, 16 Heterologous expression studies of wild-type CLCN5 in Xenopus oocytes have revealed that the channel, CLC-5, conducts chloride currents that are outwardly rectifying and time independent, and similar expression of disease associated CLC-5 mutants demonstrated markedly reduced or absent currents.8-10 To investigate further the functionally important regions of CLC-5, we pursued studies to characterize additional CLCN5 mutations in patients with Dent's disease.
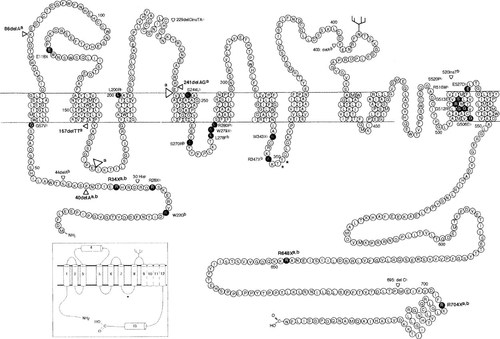
Schematic representation of CLCN5 mutations within the framework of the predicted topology of CLC-5,8 which consists of 746 amino acids.14 The correct topology of the CLC-5 putative transmembrane domains (D1-D13) is not established, and the representation is based upon the previously reported model (inset).16 The consensus phosphorylation and glycosylation sites are indicated by the asterisks and branch sites, respectively. The eight mutationsa (Arg34Stop (R34X), Arg648Stop (R648X), Arg704Stop (R704X), 40:del A, 86:del A, 157:del TT, and 241:del AG, and the loss of D3-D4 (open arrowheads) due to the acceptor splice consensus site mutation tgcag → tgaag) detected by the present study (Table 2) are indicated in bold. Of the 45 CLCN5 mutationsb reported from previous studies,8-12, 28-30 28 are shown and 6 of these have been observed to occur more than once; in addition, there have been reported 2 large deletions encompassing the entire CLCN5 gene,8, 11 2 small intragenic deletions,8, 28 and 3 splice-site mutations,8, 29 which are not shown.
MATERIALS AND METHODS
Patients
Eight probands who suffered from Dent's disease (Table 1) were investigated after obtaining informed consent. All eight of the probands had low-molecular-weight-proteinuria, seven had renal impairment, seven had nephrocalcinosis and/or nephrolithiasis, and three had rickets. Hypercalciuria was documented in seven patients, and in the remaining one (proband 4/96; Table 1) urinary calcium excretion was not measured before the onset of renal failure, which was of a severity to account for the absence of hypercalciuria. All of the eight probands were of Northern European origin. A family history of Dent's disease could be established in three probands, while in the remaining five probands, family members were not available for study to establish an inherited basis for the disease. Venous blood samples were obtained from these eight probands and three affected and nine unaffected family members for mutational analysis of the CLCN5 gene.
DNA sequence analysis of the CLCN5 gene
DNA sequence abnormalities were initially sought for using single-stranded conformational polymorphism (SSCP) analysis.10 Leukocyte DNA was extracted and used with CLCN5-specific primers10 for polymerase chain reaction (PCR) amplification utilizing conditions previously described.10 The PCR products were analyzed for abnormal SSCPs using the Phast electrophoresis system (Amersham Pharmacia, Biotech, Uppsala, Sweden). Products were run on nondenaturing polyacrylamide gels at two temperatures (10°C and 20°C),10 and the SSCP bands (Fig. 2) were revealed by silver staining with 0.025 M aqueous silver nitrate, as described.10, 17 Genomic DNA samples from unrelated normal individuals were used as controls in the SSCP analysis. The DNA sequence of PCR products with SSCP bands that differed from the normal controls was determined by Taq polymerase cycle sequencing using a semiautomated detection system (ABI 373A sequencer; Applied Biosystems, Foster City, CA).17 DNA sequence analysis of the entire 2238 bp coding region in probands in whom SSCP abnormalities were not detected was performed as described previously.10, 17 DNA sequence abnormalities were confirmed by either restriction endonuclease analysis or sequence-specific oligonucleotide (SSO) hybridization analysis of the appropriate genomic PCR products.17, 18 The DNA sequence abnormalities were demonstrated to be absent as common polymorphisms in the DNA obtained from 74 unrelated normal individuals (34 males, 40 females), and to cosegregate with the disorder in the available members from three of the families.
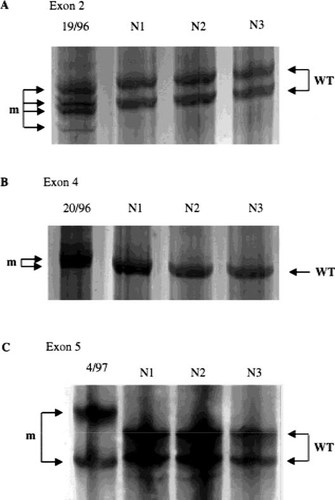
Detection of three CLCN5 mutations by SSCP in exons 2, 4, and 5. (A) shows the results of SSCP analysis of the 40delA in proband 19/96 with Dent's disease and his family (Fig. 3) together with three unrelated normals (N1 to N3). The mutation (lane 1) resulted in four mutant bands which differed from the two wild-type (WT) bands. (B) shows the results of SSCP analysis of the 157delTT in proband 20/96, together with three unrelated normals (N1 to N3). The mutation in lane 1 resulted in two mutant (m) bands which differed from the wild-type (WT) band. (C) shows the results of SSCP analysis of the 241delAG detected in the proband 4/97 together with three unrelated normals (N1 to N3). The two mutant bands (m) (lane 1) differed from both of the wild-type (WT) bands. SSCP analysis was successful in detecting six of the eight CLCN5 mutations in this study.
CLCN5 mRNA analysis
RNA was extracted from Epstein-Barr virus (EBV) transformed lymphoblastoid cell lines established from peripheral blood cells of proband 12/96 and from three unrelated normal individuals. Reverse transcriptase (RT)-PCR was performed using pairs of nested CLCN5-specific primers (outer primers-forward 5′-GTTAGCTGGTTTGATAGACA- 3′, and reverse 5′-GCCCAGCAGAATGAATG-3′; and inner primers: forward 5′-GTACGTCCTCTGGGCTCTCCTATT-3′, and reverse 5′-GATGCCATGGGGTGTGAAACTC-3′) using 25 cycles with an annealing temperature of 52°C and 57°C for the first and second rounds, respectively.8, 19 The PCR products were gel purified and the DNA sequences of both strands determined as described.8, 10, 17
RESULTS AND DISCUSSION
SSCP analysis of the entire 2238 bp coding region of the CLCN5 gene from each of the eight unrelated probands (Table 1) with Dent's disease revealed the presence of abnormal bands (Fig. 2) in the six probands designated 4.1/96, 10/96, 19/96, 20/96, 4/97, and 18/97 (Table 2). DNA sequence analysis of these SSCP abnormalities revealed the presence of six mutations (Table 2). These mutations consisted of two nonsense mutations (Arg648Stop and Arg704Stop) and four deletional mutations that involved codons 40, 86, 157, and 241 and resulted in frameshifts (Table 2 and Fig. 3). In addition, DNA sequence analysis of the entire 2238 bp coding region of the CLCN5 gene was performed in the probands designated 4/96 and 12/96, in whom SSCP abnormalities were not detected. This revealed the presence of two additional mutations; one of these was a nonsense mutation (Arg34Stop) and the other involved a c → a transversion at the –3 bp position of the acceptor splice consensus sequence of intron 5 (Table 2 and Fig. 4). The Arg34Stop, Arg648Stop, Arg704Stop, and intron 5 acceptor splice consensus sequence mutations each resulted in an alteration of a restriction enzyme site (Table 2), which facilitated their confirmation (Fig. 4). The four deletional mutations were not associated with an altered restriction enzyme site and the method of SSO hybridization analysis (Fig. 3) was used for their confirmation (Table 2). The absence of each of these eight DNA sequence abnormalities in 110 alleles from 74 unrelated normal individuals established that these abnormalities were not common sequence polymorphisms that would be expected to occur in >1% of the population.

Detection of mutation in exon 3 in family 19/96 by SSO hybridization analysis. DNA sequence analysis of the affected male II.2 revealed a 1 bp deletion (A) at codon 40. This led to a frameshift which resulted in seven missense amino acids (Ala, Lys, Ser shown) followed by a termination signal (Stop) at codon 47 (A). This deletional mutation, which did not lead to an altered restriction enzyme site, was confirmed and demonstrated to cosegregate with Dent's disease in the family by the use of SSO hybridization analysis (B). The affected males I.3, II.2, and III.3 showed hybridization only with the mutant oligonucleotide probe and were therefore hemizygous for the mutant allele, while the carrier females I.2 and I.4 were heterozygous for the mutant (m) and wild-type (WT) alleles. The absence of this 1 bp deletional mutation in 74 unrelated normal individuals (N1 to N3 shown) indicated that it was not a common DNA sequence polymorphism. Individuals are shown as: male (square), female (circle), unaffected (open), affected (filled), carrier female (dot in center of circle).
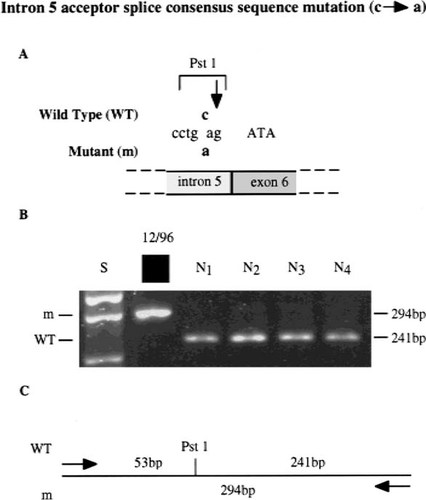
Detection of an acceptor splice consensus sequence mutation in intron 5 by restriction enzyme analysis. DNA sequence analysis of the proband 12/96 (Table 1) revealed a c → a transversion at the –3 bp position of intron 5 (A), thus altering the wild-type (WT) acceptor splice consensus sequence (tgcag) to the mutant (m) sequence (tgaag). This mutation also resulted in the loss of a Pst1 restriction enzyme site (CTGCA/G), as illustrated in the restriction map in (C). PCR amplification of exon 6 and its intron boundaries followed by Pst1 digestion (B) resulted in two products of 241 bp and 53 bp (data not shown) from the normal sequence, but only one product of 294 bp from the mutant sequence. This c → a transversion was not present in 74 normal individuals (N1 to N4 shown), thereby indicating that it was not a common DNA sequence polymorphism. The acceptor splice consensus sequence mutation was shown to result in exon skipping (Fig. 5). The standard size marker (S) in the form of the 1 kb ladder is indicated. The adjacent exon 6 sequence is shown in upper case and the intron five consensus splice sequence is shown in lower case. Similar restriction enzyme analysis was used to confirm the nonsense mutations (Arg34 Stop, Arg648Stop, and Arg704Stop) in probands 4/96, 18/97, and 10/96, respectively (Table 2).
Each of these eight mutations predicts a structurally significant alteration to the channel, CLC-5 (Table 2), and is thus likely to be of importance in the etiology of the disease. Thus, all the nonsense and deletional frameshift mutations are predicted to result in truncated CLC-5s with a loss of channel function. Indeed, two of the nonsense mutations (Arg648Stop and Arg704Stop) which have been previously observed in patient's with Dent's disease8 have been shown to result in a loss of CLC-5 channel function. However, the effects of the position −3 bp intron 5 acceptor splice consensus site mutation are more difficult to predict. The c nucleotide at position –3 of the acceptor splice consensus sequence is conserved in ∼65% of eukaryotic sequences while an a nucleotide has been observed in this position in only 4% of eukaryotic sequences.20, 21 Mutations of this –3 bp c nucleotide have been reported previously in patients with beta-thalassemia, lipoprotein lipase deficiency and cystic fibrosis.22-25 These and other studies have also revealed that mutations in acceptor splice site regions may be associated with: an accumulation of unspliced precursor mRNA, retention of incompletely spliced precursors, complete absence of transcripts, or the appearance of aberrantly processed mRNA from the use of alternative normally occuring splice sites or cryptic splice sites.24 To investigate these possibilities in the proband designated 12/96 (Fig. 4), we investigated CLCN5 mRNA processing by the detection of illegitimate transcription19 of the CLCN5 gene in EBV-transformed lymphoblastoids (Fig. 5). This revealed the presence of an aberrantly processed mRNA that was 207 bp smaller than the normal. DNA sequence analysis of the mutant CLCN5 product revealed exon skipping in which exon 6, which is 207 bp in size, was lost and exon 5 was spliced to exon 7. The loss of exon 6 removes the region encoding domains 3 and 4 (Fig. 1). Domain 3 is likely to be a transmembrane domain,26 and recent studies of domain 4 using cysteine-scanning mutagenesis indicate that domain 4, which contains a conserved motif that is essential for anion selectivity, is likely to be involved in pore formation.27 Thus, a loss of domains 3 and 4, due to exon 6 skipping, is likely to result in a loss of CLC-5 function.

Exon skipping due to an intron 5 acceptor splice consensus sequence mutation. The illegitimate transcription8, 19 of exons 5, 6, 7, and 8 of the CLCN5 gene was detected by RT-PCR using RNA obtained from EBV-transformed lymphoblastoids of normals (N1 to N3 shown) and the proband 12/96 with Dent's disease, that had the intron 5 acceptor splice consensus sequence mutation (Fig. 4). In the normal individuals, correctly spliced CLCN5 cDNA was observed (A) at the expected size of 521 bp. However, the proband with Dent's disease and the c → a transversion of the intron 5 acceptor splice site (Fig. 4) was found to have an abnormal CLCN5 cDNA of 314 bp. This mutant (m) cDNA differed from the wild-type (WT) cDNA by 207 bp, which corresponds to the size of exon 6 (B). DNA sequence analysis of the mutant cDNA confirmed exon 6 skipping, with splicing of exon 5 to exon 7. This loss of exon 6 would, if translated, result in a CLC-5 channel that lacked domains 3–4 (Fig. 1 and Table 2).
Our results, which have identified 8 CLCN5 mutations (Table 2), expand the spectrum of such mutations that are associated with Dent's disease. However, an examination of these 8 mutations together with all of the 45 previously reported CLCN5 mutations (Fig. 1),8-12, 28-30 6 of which have been reported to occur more than once, does not help to establish a correlation between the type of mutation and the phenotypic features. For example, hypophosphataemia and rickets have been observed in approximately one third of patients with Dent's disease2 and the nonsense and deletional frameshift mutations were observed in probands 4.1/96, 18/97, and 20/96 (Tables 1 and 2) that had rickets. In addition, hypophosphataemia and rickets have been observed previously in other patients with missense (Ser244Leu, Leu200Arg, Ser520Pro, Glu527Asp), insertional (30:His ins) and nonsense (Arg648Stop) mutations.8, 9 Furthermore, these mutations are not clustered but scattered throughout the coding sequence. The mechanisms whereby these CLCN5 mutations result in the diverse phenotypes of Dent's disease remain to be elucidated. However, the combined characteristics of such CLCN5 mutations together with the clinical and biochemical features in the patients with Dent's disease will help to reveal the role of CLC-5 in this renal tubulopathy that is associated with abnormal mineral metabolism.
Acknowledgements
We are grateful to the Medical Research Council (U.K.) (J.P.D. Cox, K. Yamamoto, P.T. Christie, C. Wooding, and R.V. Thakker); and to Dr. L.H. Sevitt and Prof. G.H. Nield for access to patients. J.P.D. Cox is an MRC Training Fellow.






