Lysyl Hydroxylase-2b Directs Collagen Cross-Linking Pathways in MC3T3-E1 Cells†
The authors have no conflict of interest
Abstract
To elucidate the roles of LH2b in collagen cross-linking, MC3T3-E1 cell clones expressing higher (S) or lower (AS) levels of LH2b were established. Compared with controls, the collagen cross-linking pattern was shifted toward hydroxylysine-aldehyde (S clones)- or lysine-aldehyde (AS clones)-derived pathways. The data indicate that LH2b directs collagen cross-linking pathways through its action on telopeptidyl lysine residues.
Introduction: Lysine (Lys) hydroxylation is a post-translational modification of collagen critical for cross-linking and glycosylation. Currently, three isoforms of lysyl hydroxylase (LH) have been identified, but their specific functions are still not well defined. Recently, we proposed that LH2 might modulate collagen cross-linking pattern through its action on Lys residues located in the telopeptide domains of collagen.
Materials and Methods: To directly test this hypothesis, several MC3T3-E1 cell-derived clones expressing higher (sense [S]) or lower (antisense [AS]) levels of LH2b, the predominant form of LH2 in this cell line, were established and cultured for 2 weeks, and collagen cross-links and precursor aldehydes in the matrices were analyzed.
Results: In S clones tested, the ratio of dihydroxylysinonorleucine (DHLNL) to hydroxylysinonorleucine (HLNL) was significantly higher than the average of controls (76% and 140% increase, respectively), and the level of pyridinoline (Pyr) was elevated (100% and 150% increase, respectively). In contrast, when MC3T3-E1 cells were transfected with a LH2b antisense construct (AS clones), the DHLNL/HLNL ratios were significantly lower than that of controls (56% and 73% decrease, respectively), and Pyr was not detected. Furthermore, significant amounts of an aldol-derived cross-link, dehydrohistidinohydroxymerodesmosine, were produced (∼0.3 mol/mol of collagen) in AS clones.
Conclusions: The data clearly show a critical role of LH2b in determining collagen cross-linking pathways, most likely through its action on telopeptidyl Lys residues.
INTRODUCTION
FIBRILLAR TYPE I COLLAGEN is the most abundant protein in vertebrates, providing the basis for form, support, and connectivity of the tissues and organs. One of the characteristics of collagen is its large number of post-translational modifications, many of which are unique to collagens.1 Covalent intermolecular cross-linking is the final modification essential for the stability of the fibrils, thus, for the “functional” fibrils. One of the key factors that determine the cross-linking chemistry is the state of hydroxylation of lysine (Lys) residues, which reside in the nonhelical termini of the molecule, telopeptides. Those specific residues can be converted to the respective aldehyde by the action of lysyl oxidase, and depending on the state of hydroxylation, two distinct cross-linking pathways (i.e., Lys-aldehyde [Lysald] or hydroxylysine-aldehyde [Hylald]) proceed.2-4 One of the multivalent, complex cross-links derived from the former is dehydrohistidinohydroxymerodesmosine (deH-HHMD), which involves two Lysald residues, and one of those from the latter is pyridinoline (Pyr), which involves two Hylald residues (Fig. 1).
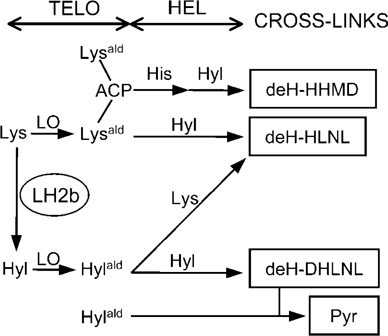
Major cross-linking pathways of collagen seen in cultures of MC3T3-E1 cells and various clones characterized in this study. LH2b, lysyl hydroxylase 2 with an alternative splice exon 13A; LO, lysyl oxidase; Lys, lysine; Hyl, hydroxylysine; His, histidine; ACP, aldol condensation product (intramolecular cross-link); Lysald, lysyl aldehyde; Hylald, hydroxylysyl aldehyde; deH, dehydro; HHMD, histidinohydroxymerodesmosine; HLNL, hydroxylysinonorleucine; DHLNL, dihydroxylysinonorleucine; Pyr, pyridinoline; TELO, nonhelical telopeptides of collagen; HEL, helical portion of collagen.
The process of Lys hydroxylation is catalyzed by lysyl hydroxylase (LH),1 and currently, three genes (procollagen-lysine, 2-oxoglutarate 5-dioxygenase, and PLOD 1-3) encoding for three isoforms of LH (LH1, LH2, and LH3) have been cloned and characterized in human,5-8 mouse,9, 10 and rat.11 More recently, an alternative splicing form of LH2, “LH2b or LH2alt,” with a 63-bp splice insert (exon 13A), has been identified.10, 12, 13 LH1 likely catalyzes Lys hydroxylation in the helical domain of collagen,14 but the specificity of LH2a, LH2b, or LH3, if any, has not been well defined as yet.
Recently, we showed that increased expression of PLOD2 mRNA coincided with an increase in Lys hydroxylation in the type I collagen telopeptides and with an increase in Hylald-derived cross-links during human osteoblastic cell differentiation. Thus, we proposed that LH2 might modulate collagen cross-linking pattern through its action on Lys residues located in the telopeptide domains of collagen.15 Here, to directly test this hypothesis, we generated several MC3T3-E1 cell-derived clones that express higher or lower levels of LH2b (the predominant form of LH2 in MC3T3-E1 cells) and investigated the effects of altered LH2b levels on collagen cross-linking in their respective cultures.
MATERIALS AND METHODS
Cell culture
MC3T3-E1 subclone 4 (referred to as MC3T3-E1 cells hereafter), a cell line that possesses a well-defined osteoblastic phenotype,16 was purchased from American Type Culture Collection (CRL-2593). The cells were grown in α-MEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Sigma, St Louis, MO, USA) and supplemented with 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B (Invitrogen).
Isolation of LH2 cDNA and generation of constructs
Total RNA was isolated from MC3T3-E1 cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The first-strand cDNA was synthesized from 2 μg of total RNA by means of the Omniscript Reverse Transcriptase Kit (Qiagen, Valencia, CA, USA). Specific primers for the coding sequence of mouse LH2 (GenBank accession NM011961) were designed using the Oligo software (Molecular Biology Insights). The sequences of the primers were as follows: upper primer, 5′GGGCGGATGGAGGACC3′ (position 3-18); and lower primer, 5′GGGATCTATAAATGACACTGCAATG3′ (position 2219-2195). This set of primers amplifies both LH2 splice variants, that is, LH2a (LH2 without splice insert) and LH2b (LH2 with splice insert). PCR amplification was performed by the ProofStart DNA polymerase (Qiagen) with an annealing temperature of 55.4°C for 35 cycles. After adding 3′A-overhangs, the PCR products were ligated into the pcDNA3.1/V5-His-TOPOvector (Invitrogen) generating the pcDNA3.1/V5-His/LH2 construct. The orientation and molecular weight of the ligated insert were analyzed by BamHI restriction enzyme digestion followed by 1.2% agarose gel electrophoresis. Plasmids containing digested inserts of the 1492 bp in the sense (S) or 792 bp in the antisense (AS) orientation were sequenced at the UNC-CH DNA Sequencing Facility. A search in the database confirmed that the ligated PCR product was 99-100% homologous to the mouse LH2 sequence previously published.9
Determination of LH2 isoform expressed by MC3T3-E1 cells
To determine whether MC3T3-E1 cells expressed LH2b and/or LH2a, specific primers were designed based on the mouse LH2 sequence and previous publications of the alternative splicing variant.10, 12 The sequences of the primers were as follows: upper primer, 5′GAAAGGAACTATTTTGTCCGTGATA3′ (position 1440-1464); lower primer, 5′GTCTGTTAGAAATGTACATAAACAC3′ (position 1575-1599). This set of primers (set I) was designed to amplify a 160-bp fragment containing the 63 bp of exon 13A in the case of LH2b and a 97-bp fragment in case of LH2a. Another upper primer (5′ATGACTTTACAAAGGGAAAAAGACT3′; position 1506-1530) was designed to anneal within the exon 13A and to be used in combination with the lower primer described above (set II). In this case, a 94-bp fragment would be amplified for LH2b, and no amplification would occur for LH2a. As positive controls for LH2a, the cDNAs from mouse heart and kidney (BD Biosciences, Palo Alto, CA, USA), tissues where LH2a mRNA is expressed,17 were used in a PCR reaction with the primer set I. All PCR reactions were performed using the HotStarTaq DNA polymerase (Qiagen) with an annealing temperature of 51.5°C for 28 cycles. The pcDNA3.1/V5-His/LH2 construct (V) was also used as a template for PCR amplification to determine the LH2 form that was cloned. All PCR products were resolved on a 12% PAGE.
Transfection and generation of stable cell clones
MC3T3-E1 cells were transfected with pcDNA3.1/V5-His/LH2 constructs (S or AS orientation) and pcDNA3.1/V5-His A vector (empty vector [EV]; Invitrogen) using FuGENE 6 transfection reagent (Roche, Indianapolis, IN, USA). After 48 h, cells were trypsinized and plated at a low density. Single cell-derived clones were isolated and maintained in the presence of 400 μg/ml G418 (Invitrogen) for up to 4 weeks.
Immunoprecipitation and Western blot analysis of the S clones
MC3T3-E1 cells, MC3T3-E1-derived cell clones transfected with the S construct, and MC3T3-E1 cells transfected with EV (EV clone) were plated onto 10-cm dishes containing α-MEM, 10% FBS, and 400 μg/ml G418. After 7 days of cell culture, cell layers/matrices were washed with PBS, detached by trypsin digestion, and counted, and same cell numbers from each cell type were lysed with a lysis buffer (150 mM NaCl, 20 mM Tris-HCl pH7.5, 10 mM EDTA, 1% Triton X-100, 1% deoxycholate, 1 mM phenylmethylsulfonylfluoride, and 1.5% aprotinin), and incubated with anti-V5 antibody (Invitrogen) at 4°C for 2 h. After addition of rec-Protein A-Sepharose 4B conjugate beads (Zymed Laboratories, South San Francisco, CA, USA) and incubation at 4°C for 30 minutes, the samples were washed with lysis buffer twice, boiled with NuPAGE LDS sample buffer (Invitrogen) under reducing conditions, resolved in a 4-12% NuPAGE Bis-Tris gel (Invitrogen), and transferred onto Immobilon-P membrane (Millipore Corp., Bedford, MA, USA). The Western blotting was performed with anti-V5 antibody, and as a secondary antibody, an anti-mouse IgG conjugated to alkaline phosphatase (Bio-Rad Laboratories, Hercules, CA, USA) was used. Immunoreactivities were visualized by alkaline phosphatase conjugate substrate kit (Bio-Rad Laboratories). Several MC3T3-E1-derived cell clones expressing higher levels of LH2b were established, and two clones (S1 and S2) expressing the highest levels of LH2b were selected for further analyses.
Verification of the AS clones by RT-PCR
Several MC3T3-E1-derived cell clones transfected with the AS construct were cultured until confluence, and total RNA extraction and first-strand cDNA synthesis was performed as described above. A set of primers that specifically amplifies a fragment of the AS transcript was designed as follows: a forward primer that anneals to the LH2 AS transcript (5′CGCCTCTCCACTCCTGAC3′) and a reverse primer that anneals to the BGH priming site of pcDNA3.1/V5-His vector (5′TAGAAGGCACAGTCGAGGCTGA3′). With this set of primers, the expected PCR product for the AS clones was 412 bp. PCR was performed by means of HotStarTaq DNA polymerase, with an annealing temperature of 57.9°C for 36 cycles. The PCR products were analyzed by 3% agarose gel electrophoresis. Two cell clones expressing the highest levels of the AS transcript (AS1 and AS2) were selected for further analyses.
Suppression of LH2b expression by transient transfection with AS construct
To verify that the AS construct suppresses the LH2b synthesis at the protein level, MC3T3-E1 cells and S1 and S2 clones were transiently transfected with this construct or EV, and the LH2b levels were examined. MC3T3-E1 cells and S1 and S2 clones were plated at a density of 2 × 105 cells, cultured overnight, and transfected with the AS construct or EV as described above. After 48 h, all samples were prepared for immunoprecipitation and Western blot analysis with anti-V5 antibodies as described above.
Expression of LH1 and LH3 mRNAs by S and AS clones
To determine if LH1 and LH3 mRNA levels were affected by changes in LH2b expression, the mRNA expression of LH1 and LH3 by S and AS clones was analyzed and compared with controls by quantitative real-time PCR. Total RNAs from MC3T3-E1 cells, EV clone, and S and AS clones cultured for 2 weeks were collected, and cDNAs were synthesized as described above. Quantitative real-time PCR analysis was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. Aliquots of each cDNA derived from 10 ng total RNA were used as templates for real-time PCR reactions with either specific primers and probe for LH1 (Applied Biosystems, assay ID; Mm00599925_m1), LH3 (assay ID; Mm00478798_m1), or GAPDH (rodent GAPDH control reagents, Applied Biosystems). All analyses were performed in triplicates. The LH1 or LH3 mRNA expression levels relative to GAPDH were analyzed using the 2−ΔΔCT method, and the fold changes were calculated using the values of MC3T3-E1 cells as a calibrator.
Collagen cross-link analysis
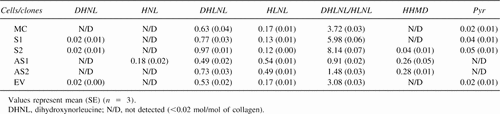
MC3T3-E1 cells, S clones (S1 and S2), AS clones (AS1 and AS2), and EV clone were cultured in α-MEM, 10% FBS, and 50 μg/ml ascorbic acid. After 2 weeks of culture, cells/matrices were scraped, thoroughly washed with PBS, and lyophilized. Two milligrams of dried samples was suspended in 0.15 M N-trismethyl-2-aminoethanesulfonic acid and 0.05 M Tris-HCl buffer (pH 7.4) and reduced with standardized NaB3H4.18 The reduced samples were hydrolyzed with 6N HCl in vacuo, after flushing with N2, at 105°C for 22 h, dried, dissolved in distilled water, and filtered. An aliquot of the hydrolysate was subjected to amino acid analysis to determine hydroxyproline (Hyp) content, and the hydrolysates with known amounts of Hyp were analyzed for cross-links on a cation-exchange column (AA-911; Transgenomic., Omaha, NE, USA) linked to a fluorescence detector (FP1520; Jasco Spectroscopic, Tokyo, Japan) and a liquid scintillation analyzer (500TR series; Packard Instrument, Meriden, CT, USA) as described previously.2 The cross-link precursor aldehydes (i.e., Hylald and Lysald), the major reducible cross-links (i.e., dehydrodihydroxylysinonorleucine/its ketoamine [deH-DHLNL], dehydrohydroxylysinonorleucine/its ketoamine [deH-HLNL]), and deH-HHMD were analyzed as their reduced forms, that is, dihydroxynorleucine (DHNL), hydroxynorleucine (HNL), DHLNL, HLNL, and HHMD, respectively. All cross-links and precursor aldehydes were quantified as moles per mole of collagen.19 Using the same lysates, the analyses were done in triplicate; the values are means ± SE.
RESULTS
Expression of LH2b in MC3T3-E1 cells
The results of PCR analysis showed that the predominant LH2 form expressed in MC3T3-E1 cells is LH2b (form with the splice insert) and that LH2a (form without the splice insert) expression in MC3T3-E1 cells was not detected (Fig. 2). Furthermore, when the pcDNA3.1/V5-His/LH2 construct (V) was used as a template for PCR amplification, a band of 160 bp (primer set I; Fig. 2A) and a band of 94 bp (primer set II; Fig. 2B) were detected, showing that the insert that was cloned and transfected into MC3T3-E1 cells was the LH2b form. When mouse heart (H) and kidney (K) were analyzed with primer set I, both LH2a (97-bp band) and LH2b (160-bp band) were clearly detected, showing the effectiveness of the primer set I to detect both LH2 forms (Fig. 2C).
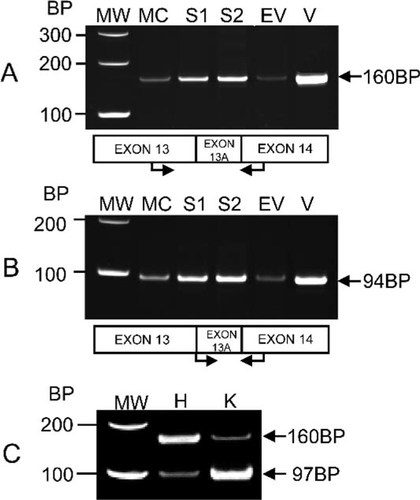
Determination of LH2 isoform expressed by MC3T3-E1 cells, S clones, and EV clone. PCR products were resolved on a 12% PAGE. (A) Detection of PCR products of 160 bp, including the 63-bp splice insert-exon 13A, amplified by primer set I (shown by arrows), indicates that the LH2b form is expressed by MC3T3-E1 cells, S clones (S1 and S2), and EV clone. (B) A 94-bp PCR product was detected with primer set II (shown by arrows), confirming the expression of LH2b by all cells/clones. (C) The 97-bp PCR product of LH2a and the 160-bp PCR product of LH2b were detected in heart and kidney, positive controls for LH2a expression, when using primer set I. MC, MC3T3-E1 cells; S1 and S2, MC3T3-E1-derived cell clones expressing higher levels of LH2b; EV, MC3T3-E1 cells transfected with an empty pcDNA3.1/V5-His A vector; V, pcDNA3.1/V5-His/LH2 construct; H, heart; K, kidney.
Generation and characterization of clones expressing higher levels of LH2
The results of the immunoprecipitation and Western blotting with anti-V5 antibodies for MC3T3-E1 cells, S clones (S1 and S2), and EV clone are shown in Fig. 3A. Immunoreactive bands at ∼97 kDa are seen only in the S clones, indicating the presence of the V5-tagged LH2b protein. The molecular weight of the immunoreactive band is consistent with that of LH2b (∼87 kDa), with an additional ∼5 kDa C-terminal peptide containing V5 epitope and a polyhistidine tag, and several N-linked glycosylation sites in LH2 that are known to occur.20 There are six potential N-linked glycosylation sites within LH2, and in fact, the carbohydrate moieties seem to be required for optimal LH activity.20, 21 No immunopositive band was detected in MC3T3-E1 cells or the EV clone.

Detection of (A and C) V5-tagged LH2b protein and (B) LH2b AS transcript by immunoprecipitation-Western blot analysis using anti-V5 antibodies and RT-PCR using specific primers as described in the Materials and Methods section, respectively. (A) The immunoreactive bands at ∼97 kDa were present only in the S clones (S1 and S2). The ∼50-kDa band seen in each sample is the heavy chain of IgG. (B) The RT-PCR bands of 412 bp were seen only in the AS clones (AS1 and AS2), indicating the presence of LH2 AS transcript in these clones. (C) The immunoreactive bands corresponding to the V5-tagged LH2b protein were significantly reduced when S clones were transfected with the LH2 AS construct but not with the EV. AS1 and AS2, MC3T3-E1-derived cell clones transfected with a LH2b AS construct.
Detection of the AS transcript and suppression of LH2 protein by LH2 AS construct
The RT-PCR result of MC3T3-E1 cells, AS clones (AS1 and AS2), and EV clone using primers designed to specifically amplify a fragment of the LH2 cDNA in an antisense orientation are shown in Fig. 3B. The expected bands of 412 bp are seen only in the AS clones, indicating the presence of LH2 AS transcript in these clones. To verify that the AS transcript suppresses the LH2b synthesis at the protein level, MC3T3-E1 cells and S1 and S2 clones were transiently transfected with the AS construct, and the inhibition of LH2b synthesis was analyzed as described in the Materials and Methods section. As shown in Fig. 3C, V5-tagged LH2b protein levels in S1 and S2 clones were clearly inhibited by the transfection with the LH2 AS construct, indicating that the AS clones established in this study reduced the endogenous LH2b the protein level.
Expression of LH1 and LH3 mRNAs by S and AS clones
The expression of LH1 and LH3 mRNAs by S (S1 and S2) and AS clones (AS1 and AS2) was analyzed by quantitative real-time PCR, and they were essentially unchanged in both S and AS clones compared with those of controls. Three independent experiments produced similar results, and a representative result is shown in Fig. 4.
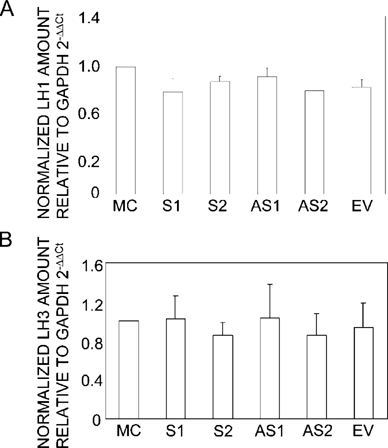
Expression of LH1 and LH3 mRNAs in MC3T3-E1 cells, S clones (S1 and S2), AS clones (AS1 and AS2), and EV clone analyzed by quantitative real-time PCR. The LH1 or LH3 mRNA expression levels relative to GAPDH were analyzed using the 2−ΔΔCT method, and the fold changes were calculated using the values of MC3T3-E1 cells as a calibrator. The values represent normalized (A) LH1 or (B) LH3 mRNA levels and are shown as the mean ± SD based on triplicate assays.
Collagen cross-link analysis
The collagen cross-links produced in MC3T3-E1 cells, EV clone, and S clones (S1 and S2) at 2 weeks of culture were bifunctional reducible cross-links (i.e., DHLNL and HLNL, as major cross-links and Pyr as a minor one). In the case of AS clones (AS1 and AS2), in addition to DHLNL and HLNL, a tetravalent, aldol-derived reducible cross-link, HHMD, was found as another major cross-link, whereas no Pyr was detected. Deoxypyridinoline was not detected in any of the hydrolysates. The chromatographs for reducible cross-links obtained from the controls (MC3T3-E1 cells and EV clone), S clones, and AS clones are shown in Fig. 5. The actual concentrations of DHLNL, HLNL, HHMD, Pyr cross-links, precursor aldehydes (DHNL and HNL), and the ratio of DHLNL to HLNL are depicted in Table 1. The ratio of DHLNL to HLNL in MC3T3-E1 cells is similar to that previously reported.22 The most striking differences among the S clones, AS clones, and the control groups were the ratios of DHLNL to HLNL and the levels of Pyr and HHMD. The ratio in two control groups, MC3T3-E1 cells and EV clone, was comparable, and it was ∼3.7 and 3.1, respectively. However, it was markedly higher in S clones (6.0 and 8.1), and significantly lower in AS clones (only 0.9 and 1.5). The increase in DHLNL to HLNL ratio in S clones was proportional to the levels of LH2b in the respective clones. The high ratio seen in the S clones was apparently caused by increases in DHLNL and decreases in HLNL. The low ratio in the AS clones was mainly caused by the increase in HLNL. The Pyr contents were constantly higher in S clones (100% and 150% increase) than those of controls, whereas Pyr was not detected in the AS clones. Furthermore, significant amounts of an aldol-derived tetravalent cross-link, HHMD, were found in the AS clones (∼0.3 mol/mol of collagen), whereas it was minimal in both S clones and controls. In the AS1 clone, a significant amount of HNL (reduced Lysald) was found (0.2 mol/mol of collagen). The cross-linking pattern of EV clone was essentially identical to that of MC3T3-E1 cells.
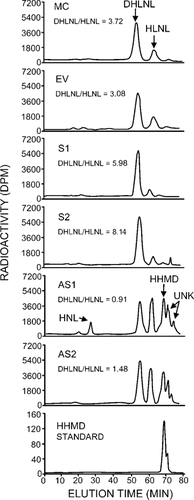
Reducible cross-link pattern of collagen obtained from MC3T3-E1, EV clone, S clones, and AS clones at 2 weeks of cell culture. The reduced collagen hydrolysates containing 300 nmol of Hyp were analyzed by HPLC. Note the increase in DHLNL to HLNL ratio in S clones and the decrease in this ratio in AS clones compared with that of the control groups (MC3T3-E1 and EV). Significant amounts of HHMD and HNL were found only in AS1 clone. HNL, hydroxynorleucine or reduced Lysald; UNK, unidentified peaks.
DISCUSSION
The key to cross-linking chemistry of type I collagen is the post-translational chemical state (hydroxylation) of five Lys residues in the telopeptides that can be converted to aldehyde. The aldehyde produced initiates a series of condensation reactions forming intra- and intermolecular covalent cross-links.19 It has been long speculated that Lys residues in the telopeptides are hydroxylated by mechanisms different from those in the helical domain of the molecule (e.g., through different LHs).23 Recent findings of several LH isoforms led us to pursue the possibility that one of the isoforms might function specifically as a telopeptidyl LH. In our previous studies on human osteoblastic cell differentiation and matrix mineralization, we found that the expression pattern of PLOD2, which encodes for LH2, was well correlated with the telopeptidyl Lys hydroxylation and subsequent cross-linking pattern, suggesting the role of LH2 as a telopeptidyl LH.15 Most recently, Mercer et al.17 reported that transfection of LH2 into CHO-K1 cells leads to an increase in Pyr cross-links, indicating that LH2 possesses telopeptidyl LH activity. Furthermore, mutation analysis in patients with Bruck syndrome, which is characterized by Pyr deficiency in bone collagen, showed two missense mutations in exon 17 of PLOD2,24 implying LH2 as a putative telopeptidyl LH.
In this study, to obtain more direct insights into the roles of LH2 in collagen cross-linking, we have established several MC3T3-E1-derived clones expressing higher or lower levels of LH2b (S and AS clones, respectively) and characterized cross-linking pattern of collagen produced by these clones. In MC3T3-E1 cells, it was found that the alternative splice variant LH2b, containing a 21 amino acid proline-rich sequence, is the predominant form of LH2. This is consistent with a previous report showing that LH2b is the major isoform in several cells and tissues.12 When LH2b was overexpressed in MC3T3-E1 cells, significant changes occurred in collagen cross-linking, that is, increased DHLNL/HLNL ratio (caused by an increase in DHLNL and a concomitant decrease in HLNL) and increased Pyr, showing an increase in the extent of Lys hydroxylation in collagen cross-links. Although the increased DHLNL (Hylald × Hyl)/HLNL (Lysald × Hyl or Hylald × Lys; Fig. 1) could be caused by an increase in Lys hydroxylation in the helical counterpart with the same level of telopeptidyl Lys hydroxylation, the increase is most likely caused by an increase in Lys hydroxylation in the telopeptides because (1) the mRNA expression level of LH1, a helical LH, was unchanged in these clones; (2) a significant increase in Pyr (Hylald × Hylald × Hyl) seen in the S clones is clear evidence for elevated Lys hydroxylation in the telopeptides; and (3) our previous study on human osteoblastic cells clearly showed a correlation of LH2 expression and telopeptide Lys hydroxylation.15 Two recent reports17, 24 also support the above notion. Furthermore, in AS clones, not only the ratio of DHLNL to HLNL decreased, but also significant amounts of an aldol-derived cross-link, HHMD (Lysald × Lysald × His × Hyl), was produced (Fig. 5). This cross-link is generally minimal in quantity in collagen from mineralized tissues,25, 26 as well as in collagen produced by osteoblastic cells22 (Table 1). In addition, Pyr was not detected in the AS clones. Thus, S and AS clones lead to two distinct collagen cross-linking pathways that stem from the state of telopeptidyl Lys hydroxylation. To the best of our knowledge, this represents the first report showing that collagen cross-linking pathways can be switched by manipulating the specific LH expression.
In conclusion, it was shown that manipulation of LH2b expression results in changes in collagen cross-linking, likely through its action on the telopeptidyl Lys residues. Thus, LH2b regulates the pattern of collagen cross-linking, and its differential expression may partially explain the well-known tissue-specific collagen cross-linking pattern.4, 27, 28
Acknowledgements
This study was supported by NASA Grant NAG2-1596 and NIH Grant DE10489.