Molecular Characterization and Genetic Diversity of Isolated Foot-and-Mouth Disease Viruses Circulating in Cattle in The Mekong Delta Provinces, Vietnam
Abstract
Foot-and-mouth disease (FMD) is a highly contagious viral disease of cloven-footed livestock caused by foot-and-mouth disease virus (FMDV). FMD has significant impacts on farmers and national economies. The evolution and mutation of FMDV have contributed to the emergence of new strains of FMDV. Sequences of VP1 from 11 FMDV isolates in the Mekong Delta Provinces were obtained by Sanger sequencing technology. The phylogenetic analysis of VP1 sequence elucidated that 8 FMDV isolates including O-VN-CTU-VL01 (PP897837), O-VN-CTU-VL02 (PP897838), O-VN-CTU-TV01 (PP897840), O-VN-CTU-TV02 (PP897841), O-VN-CTU-TV03 (PP897842), O-VN-CTU-TV04 (PP897844), O-VN-CTU-BT04 (PP897847), and O-VN-CTU-BT05 (PP897847) were clustered into Group 1. On the other hand, 3 FMDV isolates including O-VN-CTU-BT01 (PP897844), O-VN-CTU-BT02 (PP897844), and O-VN-CTU-BT03 (PP897844) were clustered into Group 2. In addition, the nucleotide and deduced amino acid sequences of VP1 in Group 1 were closely related to lineage Mya-98, topotype SEA, and Type O (89%–93% nucleotide identity and 91%–99% amino acid identity). The similarity of FMDV isolates in Group 2 was closely related to lineage Pan Asia, topotype ME-SA, Type O (91.13%–94.53% and 96%–99.44% nucleotide and amino acid similarities, respectively). Analysis of amino acid sequences of VP1 illustrated several substitution mutations detected at amino acid positions 133–158 (the main antigenic site) in lineages Mya-98 (O/SEA) and Pan Asia, (O/ME-SA). Notably, a substitution mutation at position M144V was detected in FMDVs O-VN-CTU-VL1 (PP897837) and FMDV O-VN-CTU-TV1 (PP897840). No recombinant events were detected at VP1 sequences. In brief, genetic analysis of VP1 nucleotide and amino acid sequences of isolated FMDVs contributed to detecting the mutation which was able to cause the emergence of new strains as well as to elucidate the evolution of FMDVs circulating in the Mekong Delta Provinces.
1. Introduction
Foot-and-mouth disease (FMD) is a highly contagious disease of cloven-hoofed animals. The disease was initially described in the 16th century and was the first animal pathogen identified as a virus [1]. FMD is classified in List A of OIE-listed diseases because it is capable of rapidly infecting and spreading within and between countries, resulting in great economic losses [2]. The causative agent of FMD is foot-and-mouth disease virus (FMDV), a member of the genus Aphthovirus in the family Picornaviridae, which is a nonenveloped, icosahedral virus 26–30 nm in diameter containing a single positive-sense RNA genome approximately 8300 nucleotides in length [3]. The FMDV genome encodes a single polyprotein composed of four structural proteins (VP1–VP4) and ten nonstructural proteins (L, 2A-2C, 3A, 3B1–3B3, 3C, and 3D). Four structural proteins (VP1–4) contribute to the formation of the complete viral capsid. The structural proteins VP1, VP2, and VP3 are located on the surface of the virus, while VP4 is located on the inside of the virus. Among viral proteins, VP1 is closely related to viral attachment, protective immunity, and serotype specificity [4].
FMDV has a high degree of genetic and antigenic variation, which accounts for the existence of seven main FMDV serotypes, namely, O, A, C, Asia 1, Southern African Territories (SAT) 1, SAT 2, and SAT 3 [5, 6] and many subtypes [7]. FMDV Serotype O is classified into 11 topotypes, designated as Europe–South America (Euro–SA), Middle East–South Asia (ME–SA), Southeast Asia (SEA), Cathay (CHY), West Africa (WA), East Africa 1 (EA-1), East Africa 2 (EA-2), East Africa 3 (EA-3), East Africa 4 (EA-4), Indonesia-1 (ISA-1), and Indonesia-2 (ISA-2) [8, 9]. In addition, some serotypes have a restricted geographical distribution, e.g., Asia-1, whereas others, notably Serotype O, occur in many different regions. There is no cross-protection between serotypes and sometimes protection conferred by vaccines even of the same serotype can be limited [10].
In Vietnam, the FMDV Serotype O and its outbreak were first reported in academic research between 1996 and 2001 [11]. In addition, the FMDV Serotypes Asia 1 and A were subsequently identified in 2005 and 2009, respectively [12, 13]. In 2008 and 2009, FMDV Serotypes O and A were reported to be the prevalent serotype and caused almost FMD outbreaks. Then, the first detection of FMDV O/Ind-2001d in 2017 was reported in [14]. Moreover, cattle are commonly vaccinated against FMDV Serotype O twice annually with varying vaccine formulations, based upon current knowledge of circulating strains. However, sporadic FMD outbreaks have been reported in the Mekong Delta Provinces. Prominent factors of the disease are poor cross-protection between serotypes, rapid spread, and the diversity of serotypes. Currently, limited information is available regarding the genetic characteristics and geographical distribution of the FMDV causing sporadic outbreaks in the Mekong Delta Provinces. In this study, the VP1 nucleotide and amino acid sequences of 11 FMDV isolates collected from endemic outbreaks in some provinces in the Mekong Delta Provinces (Southern Vietnam) during 2022–2023 were analyzed to investigate their phylogeny and genetic relationship with other published FMDVs.
2. Materials and Methods
2.1. Ethical Approval
All experimental protocols were approved by the Institutional Animal Care and Use Committee of Can Tho University, Vietnam.
2.2. Sample Collection
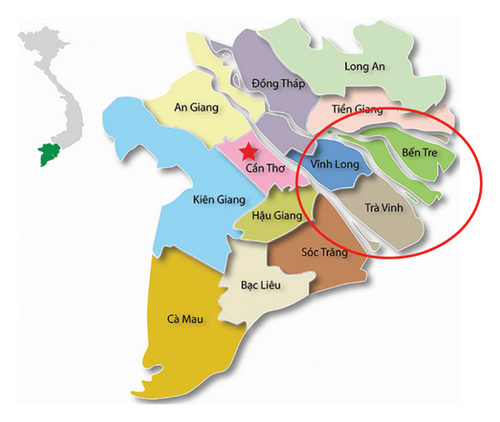
The samples were collected and processed following the World Organization for Animal Health (OIE) standard criteria [15, 16]. A total of 40 samples (damaged skin, vesicles, and/or probang) were collected from cattle exhibiting symptoms of FMD (Table 1) in FMD outbreaks located in the Mekong Delta Provinces consisting of Ben Tre, Tra Vinh, and Vinh Long Provinces. The location of those provinces is shown in Figure 1. The collected samples were stored in ice bins on the way to the laboratory and stored in the freezer (−70°C).
| No. | Accession no. | Strain | Lineage | Topotype | Country | Year | Host |
|---|---|---|---|---|---|---|---|
| 1 | PP897837 | O-VN-CTU-VL01 | Vietnam: Vinh Long | 2023 | Cattle | ||
| 2 | PP897838 | O-VN-CTU-VL02 | Vietnam: Vinh Long | 2023 | Cattle | ||
| 3 | PP897840 | O-VN-CTU-TV01 | Vietnam: Tra Vinh | 2023 | Cattle | ||
| 4 | PP897841 | O-VN-CTU-TV02 | Vietnam: Tra Vinh | 2023 | Cattle | ||
| 5 | PP897842 | O-VN-CTU-TV03 | Vietnam: Tra Vinh | 2023 | Cattle | ||
| 6 | PP897843 | O-VN-CTU-TV04 | Vietnam: Tra Vinh | 2023 | Cattle | ||
| 7 | PP897844 | O-VN-CTU-BT01 | Vietnam: Ben Tre | 2023 | Cattle | ||
| 8 | PP897845 | O-VN-CTU-BT02 | Vietnam: Ben Tre | 2023 | Cattle | ||
| 9 | PP897846 | O-VN-CTU-BT03 | Vietnam: Ben Tre | 2023 | Cattle | ||
| 10 | PP897847 | O-VN-CTU-BT04 | Vietnam: Ben Tre | 2022 | Cattle | ||
| 11 | PP897848 | O-VN-CTU-BT05 | Vietnam: Ben Tre | 2022 | Cattle | ||
| 12 | HQ260718.1 | O/VN/YB08/2010 | Mya-98 | SEA | Vietnam: Yen Bai | 2010 | Cattle |
| 13 | KC438373.1 | SKR/12/2010 | Mya-98 | SEA | South Korea | 2010 | Cattle |
| 14 | MH791315.1 | Mya-98 | Mya-98 | SEA | China | 2018 | Cattle |
| 15 | HQ116278 | O/VIT/4/2005 | Mya-98 | SEA | Vietnam | 2005 | Cattle |
| 16 | JQ070319.1 | MYA/3/2010 | Mya-98 | SEA | Myanmar | 2010 | Cattle |
| 17 | HQ116290 | O/VIT/7/2006 | Mya-98 | SEA | Vietnam | 2006 | Cattle |
| 18 | HQ116256.1 | O/TAI/1/2009 | Mya-98 | SEA | Thailand | 2009 | Cattle |
| 19 | KT153102.1 | O/VIT/128/2009 | Mya-98 | SEA | Vietnam: Ha Giang | 2009 | Cattle |
| 20 | HQ260716.1 | O/VN/DB04/2010 | Mya-98 | SEA | Vietnam: Dien Bien | 2010 | Cattle |
| 21 | JQ070325.1 | MOG/CO3/2010 | Mya-98 | SEA | Mongolia | 2010 | Cattle |
| 22 | KT153149.1 | O/VIT/40/2009 | Mya-98 | SEA | Vietnam: Ha Tinh | 2009 | Cattle |
| 23 | AJ294906.1 | O/CAM/11/94 | Cam-94 | SEA | Cambodia | 1998 | Cattle |
| 24 | AJ294929.1 | O/VIT/2/97 | Cam-94 | SEA | Vietnam | 1997 | Cattle |
| 25 | AJ303481.1 | O ALG/1/99 | WA | Algeria | 1999 | — | |
| 26 | AJ303488.1 | O GHA/5/93 | WA | Ghana | 1993 | — | |
| 27 | AJ303511.1 | KEN/83/79 | EA | Kenya | 1979 | — | |
| 28 | AJ303514.1 | O/KEN/2/95 | EA | Kenya | 1995 | — | |
| 29 | KT153137.1 | O/VIT/30/2012 | Pan Asia | ME-SA | Vietnam: Son La | 2012 | Cattle |
| 30 | KT153143.1 | O/VIT/362/2012 | Pan Asia | ME-SA | Vietnam: Long An | 2012 | Cattle |
| 31 | HQ116173.1 | O/CAM/5/2006 | Pan Asia | ME-SA | Cambodia | 2006 | Cattle |
| 32 | HQ116283.1 | O/VIT/17/2005 | Pan Asia | ME-SA | Vietnam | 2005 | Cattle |
| 33 | HQ116174.1 | O/CAM/1/2008 | Pan Asia | ME-SA | Cambodia | 2008 | Cattle |
| 34 | HQ116274.1 | O/VIT/7/2004 | Pan Asia | ME-SA | Vietnam | 2004 | Cattle |
| 35 | KM921876.1 | UAE/4/2008 | Ind2001c | ISA | United Arab Emirates | 2008 | Gazelle |
| 36 | DQ164864.1 | O/BHU/7/2002 | Ind2001a | ISA | Bhutan | 2002 | |
| 37 | DQ164969.1 | O/SAU/3/2001 | Ind2001a | ISA | Saudi Arabia | 2001 | Cattle |
| 38 | DQ164863.1 | O/BAR/1/2001 | Ind2001a | ISA | Bahrain | 2001 | Cattle |
| 39 | KC506543.1 | O/IND62/2011 | Ind2001a | ISA | India: Maharashtra | 2001 | Cattle |
| 40 | HQ630676.1 | BAN/1/2009 | Ind2001b | ISA | Bangladesh | 2009 | Cattle |
| 41 | KC506512.1 | O/IND34/2012 | Ind2001b | ISA | India: Karnataka | 2011 | Cattle |
| 42 | HQ630694.1 | NEP/2/2009 | Ind2001b | ISA | Nepal: Kathmandu | 2009 | Cattle |
| 43 | KC506471.1 | O/IND129/2010 | Ind2001b | ISA | India: Assam | 2010 | Cattle |
| 44 | MT936117.1 | IC491/2018- | Ind2001e | ISA | India: Karnataka | 2018 | Cattle |
| 45 | KY399464.1 | O/VIT/9595/2015 | Ind2001d | ISA | Vietnam: Dak Nong | 2015 | Cattle |
| 46 | KY399468.1 | O/VIT/19350/2015 | Ind2001d | ISA | Vietnam: Ninh Thuan | 2015 | Cattle |
| 47 | KJ825807.1 | IND26(54)/2014 | Ind2001d | ISA | India: Assam | 2014 | Cattle |
| 48 | KT037119.1 | BD Gh 2/2013 | Ind2001d | ISA | Bangladesh | 2013 | Cattle |
| 49 | KM921865.1 | NEP/6/2014 | Ind2001d | ISA | Nepal: Ilam | 2014 | Cattle |
| 50 | K01201.1 | O/Campos/Brazil/71 | — | EURO-SA | Brazil | 1971 | — |
| 51 | M55287.1 | O/Italy/Brescia/1947 | — | EURO-SA | Italy | 1947 | — |
| 52 | AF154271.1 | Tau-YuanTW97 | — | CATHAY | Taiwan | 1997 | Pig |
| 53 | AJ296321.1 | O/TAW/81/97 | — | CATHAY | Taiwan | 1997 | Pig |
| 54 | AY317098.1 | O/HKN/2002 | — | CATHAY | China | 2002 | Pig |
| 55 | DQ165025.1 | O/VIT/13/2002 | — | CATHAY | Vietnam | 2002 | Pig |
2.3. Primer Design
Primer design was performed as described previously in [17]. Two sets of gene-specific primers shown in Table 2 are designed using NCBI Primer3 (available on the website: https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and BLAST tools (available on the website: https://blast.ncbi.nlm.nih.gov). Then, the specificity of the designed primers was evaluated based on the aligning primers with reference FMDV strain O/YS/CHA/05 (HM008917) using MEGA 6.06 software.
| No. | Primers | Sequence 5′-3′ | Location | Product size (bp) |
|---|---|---|---|---|
| Real-time RT-PCR, UTR [18] | ||||
| 1 | FP | GTA AAC CAC TGG TTG GCT GAC T | 56 | |
| RP | TAC TCG CCG TGG CCT CG | |||
| RT-PCR, VP1 | ||||
| 2 | FP | GAC TTY GAG YTR CGB YTR CC | 3374–3394 | 744 |
| RP | GGG TTG GAC TCV ACG TCY CC | 4118–4088 | ||
2.4. FMD Screening Using Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.4.1. RNA Extraction
Total RNA was extracted from collected samples using an RNA extraction kit (NEXprep™, Korea) as described by the manufacturer’s instructions.
2.4.2. cDNA Synthesis
First-strand cDNA was synthesized using a Sensi FAST™ cDNA synthesis kit (Bioline, UK). In brief, a 20-μL total reaction volume including 10 μL of RNase-free water, 5 μL of extracted RNA sample, 1 μL of reverse transcriptase primer, and 4 μL of 5x TransAmp Buffer was prepared in 1.5-mL Eppendorf tube and kept on ice. Then, the reaction was incubated at 25°C for 10 min followed by heating at 45°C for 10 min and 85°C for 5 min and chilled on ice for at least 1 min. The synthesis cDNA was immediately used for real-time PCR or PCR amplification.
2.4.3. Real-Time PCR
For the detection of FMDV, primers directed toward the conserved UTR are used as previously described in [18] (Table 2). 2x SensiFAST™ SYBR (Bioline, UK) was used for UTR amplification, following the manufacturer’s instructions. The PCR master mix was prepared in a total volume of 15 μL. It consisted of 3.4 μL of dH2O, 10 μL of 2x SensiFAST™ SYBR, and 0.8 μL of each primer (10 pmol/μL). Template cDNA was added at 5 μL for a complete volume of 20 μL. The mixture was placed in FQD-96A (Bioer, USA) with the following cycling conditions: initial denaturation (95°C for 120 s) and 40 cycles of amplification (95°C for 5 s, 60°C for 10 s, and 72°C for 15 s) and ending with a melting curve from 60°C to 90°C.
2.5. RT-PCR
RT-PCR was carried out to amplify VP1 of FMDVs. The forward and reverse primers used in this assay are shown in Table 2.
The PCR was performed by using a MyTaq™ DNA Polymerase kit (Bioline, UK) according to the manufacturer’s instructions. In brief, 20 μL of master mix including 6.5 μL of nuclease-free water, 12.5 μL of MyTaq Mix 2X, and 1 μL of 20 μM forward and reverse primers were combined with 5 μL of cDNA to reach a total of 25 μL reaction. Subsequently, PCRs were performed following the protocol: 1 cycle of initial denaturation at 95°C for 15 s, a sequence of 35 cycles followed by denaturation at 95°C for 15 s, annealing at 60°C for 15 s, extension at 72°C for 30 s, and 1 cycle of final extension 72°C for 3 min. The PCR products were detected on 1%, 5% (w/v) agarose gel electrophoresis, and the gel picture was captured by using a UV light transilluminator (UVDI, USA).
2.6. Sequencing and Phylogenetic Analysis
Sequencing was performed in both directions with virus-specific primers. Sequences were analyzed using the Genetyx Version 12. This program was used to read the sequencing electropherograms to exclude nucleotide ambiguity. Sequences of FMDV trains were aligned with the ClustalW multiple alignment method using BioEdit 7.2.0 [19, 20]. Phylogenetic analyses were conducted using MEGA 6.06 [21], and phylogenetic trees were constructed based on strains in various geographic origins and topotypes (strains and accession numbers were retrieved from GenBank) by using the maximum likelihood method based on the Tamura Parameter 3 model with 1000 bootstrap replicates.
2.7. Recombination Analysis
Recombination analysis was performed as described previously in [17]. Briefly, eleven FMDV strains and reference FMDVs were performed for recombination analysis by using Simplot v3.5 and recombinant detection program v4.70 (RDP4). Both programs were applied to identify putative recombination breakpoints and parental isolates of recombinants. The Simplot program is performed based on a sliding window method. Nucleotide identity was calculated using the Kimura 2-parameter method with a transition–transversion ratio of 2, and the window width and step size, respectively, were 200 bp and 20 bp. Bootscan analysis was also carried out using a signal of 70% of the observed permuted tree. In the RDP4 program, various potential recombination methods including RDP, Geneconv, Bootscan, MaxChi, Chimaera, Siscan, and Topal DSS were used to identify the recombination events and parental isolates of recombinants, with significance set at p values < 0.05.
3. Results
3.1. FMDV Screening Using Real-Time RT-PCR
Out of 40 collected samples, 11 samples were positive with FMDV accounting for 27.50%. Positive samples had Ct values ranging from 15.45 to 25.59 (Table 3). During the sample collection, the FMDV-positive cattle observed salivation, pyrexia, and vesicle formations in the buccal and nasal mucous membranes and/or between the claws, and lameness.
| No. | Accession no. | Isolates | Origin | Cycle threshold | Clinical signs | Type of sample |
|---|---|---|---|---|---|---|
| 1 | PP897837 | O-VN-CTU-VL01 | Vinh Long | 23.60 | Pyrexia (> 41°C), blisters in the buccal and nasal mucous membranes | Damaged skin and vesicles |
| 2 | PP897838 | O-VN-CTU-VL02 | Vinh Long | 21.69 | Pyrexia (> 41°C), blisters in the buccal and nasal mucous membranes | Damaged skin and vesicles |
| 3 | PP897840 | O-VN-CTU-TV01 | Tra Vinh | 24.74 | Pyrexia (> 41°C), drooling | Probang |
| 4 | PP897841 | O-VN-CTU-TV02 | Tra Vinh | 15.45 | Blisters in the buccal and nasal mucous membranes and between the claws, lameness | Damaged skin and vesicles |
| 5 | PP897842 | O-VN-CTU-TV03 | Tra Vinh | 22.64 | Pyrexia (> 41°C), drooling | Probang |
| 6 | PP897843 | O-VN-CTU-TV04 | Tra Vinh | 23.38 | Blisters between the claws, lameness | Damaged skin and vesicles |
| 7 | PP897844 | O-VN-CTU-BT01 | Ben Tre | 23.01 | Pyrexia (> 41°C), blisters in the buccal and nasal mucous membranes | Damaged skin and vesicles |
| 8 | PP897845 | O-VN-CTU-BT02 | Ben Tre | 24.90 | Pyrexia (> 41°C), blisters between the claws, lameness | Damaged skin and vesicles |
| 9 | PP897846 | O-VN-CTU-BT03 | Ben Tre | 25.12 | Pyrexia (> 41°C), blisters between the claws, lameness | Damaged skin and vesicles |
| 10 | PP897847 | O-VN-CTU-BT04 | Ben Tre | 25.59 | Pyrexia (> 41°C), drooling | Probang |
| 11 | PP897848 | O-VN-CTU-BT05 | Ben Tre | 21.20 | Pyrexia (> 41°C), drooling | Probang |
3.2. Phylogenetic Analysis and Pairwise Sequence Comparison Analysis (PSCA) of VP1
In the present study, the phylogenetic relationship between the sequences of 11 FMDV isolates and 44 published FMDVs was assessed based on the VP1 sequence at Positions 1–633. The results showed that isolates O-VN-CTU-VL01 (PP897837), O-VN-CTU-VL02 (PP897838), O-VN-CTU-TV01 (PP897840), O-VN-CTU-TV02 (PP897841), O-VN-CTU-TV03 (PP897842), O-VN-CTU-TV04 (PP897844), O-VN-CTU-BT04 (PP897847), and O-VN-CTU-BT05 (PP897847) clustered into Group 1 (90.33%–97.96% nucleotide identity and 94.12%–99.44% amino acid identity) while isolates O-VN-CTU-BT01 (PP897844), O-VN-CTU-BT02 (PP897844), and O-VN-CTU-BT03 (PP897844) clustered into Group 2 (92.65%–95.82% nucleotide identity and 96.60%–98.88% amino acid identity). According to the phylogenetic tree (Figure 2), isolates in Group 1 had a close relationship to lineage Mya-98, topotype SEA, and Type O (O/SEA). In addition, isolates in Group 2 shared a close relationship to lineage Pan Asia, topotype ME-SA, and Type O (O/ME-SA).
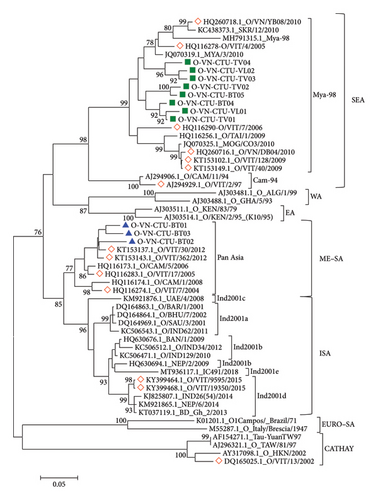
PSCA of VP1 nucleotide and amino acid sequences showed that Group 1 shared high identity with those strains in lineage Mya-98 (O/SEA) including O/VIT/4/2005 (HQ116278, Vietnam), MYA/3/2010 (JQ070319.1, Myanmar), and SKR/12/2010 (KC438373.1, South Korea) with approximately 89%–93% nucleotide identity and 91%–99% amino acid identity. Those reference strains were reported in different parts of Northern Vietnam and Asian countries. Besides, those strains in Group 1 shared 87%–90% nucleotide identity and 92%–97% amino acid identity with MOG/CO3/2010 (JQ070325.1, Mongolia) and O/TAI/1/2009 (HQ116256.1, Thailand). Meanwhile, Group 2, respectively, shared 91.13%–94.53% and 96%–99.44% nucleotide and amino acid similarities with those strains in lineage Pan Asia (O/ME-SA) consisting of O/VIT/7/2004 (HQ116274.1, Vietnam) and O/CAM/5/2006 (HQ116173.1, Cambodia). In addition, phylogenetic and PSCA of VP1 sequences showed that all FMDV isolates distinctly separated from the strains in topotypes CHY, WA, EA, EURO–SA, and ISA with approximately 73%–90% nucleotide similarity and 83%–97% amino acid similarity (Table 4).
| Amino acid similarity (%) | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
| 1 | 97.14 | 98.28 | 95.31 | 96.56 | 96.55 | 92.13 | 91.54 | 93.37 | 97.12 | 94.70 | 93.43 | 93.40 | 94.06 | 92.79 | 95.90 | 95.84 | 95.88 | 86.10 | 92.92 | 91.00 | 86.07 | 90.83 | 87.56 | 92.74 | 92.23 | 92.90 | 92.84 | |
| 2 | 91.69 | 95.91 | 95.32 | 99.44 | 99.44 | 94.05 | 93.47 | 95.26 | 97.13 | 94.72 | 95.31 | 95.28 | 95.98 | 95.90 | 98.86 | 97.13 | 98.26 | 87.54 | 95.97 | 94.15 | 88.73 | 92.79 | 90.88 | 94.11 | 93.58 | 94.76 | 94.19 | |
| 3 | 97.96 | 93.03 | 94.65 | 95.32 | 96.52 | 90.07 | 89.47 | 91.36 | 97.69 | 94.04 | 91.44 | 91.39 | 93.39 | 91.43 | 94.63 | 95.18 | 94.60 | 84.26 | 91.57 | 89.60 | 85.68 | 89.21 | 86.05 | 90.71 | 90.20 | 90.90 | 90.83 | |
| 4 | 92.72 | 91.13 | 92.89 | 94.72 | 94.71 | 89.92 | 88.83 | 90.72 | 97.11 | 99.43 | 90.81 | 90.75 | 92.77 | 92.11 | 94.02 | 94.57 | 94.67 | 84.47 | 90.95 | 88.96 | 85.55 | 88.08 | 86.05 | 90.07 | 89.56 | 90.27 | 90.20 | |
| 5 | 90.73 | 97.78 | 92.09 | 90.73 | 98.87 | 94.10 | 93.53 | 95.31 | 96.54 | 94.12 | 95.35 | 95.33 | 95.39 | 95.30 | 98.28 | 96.54 | 97.68 | 87.65 | 95.39 | 93.55 | 88.78 | 92.86 | 90.96 | 94.17 | 93.64 | 94.17 | 94.25 | |
| 6 | 90.93 | 95.10 | 92.28 | 91.12 | 95.29 | 93.42 | 92.84 | 94.64 | 97.71 | 94.10 | 94.69 | 94.66 | 95.38 | 95.29 | 98.28 | 96.53 | 97.67 | 87.31 | 95.37 | 93.53 | 89.45 | 92.64 | 90.22 | 93.49 | 92.96 | 94.15 | 93.58 | |
| 7 | 80.58 | 83.56 | 80.87 | 80.15 | 82.42 | 81.54 | 96.60 | 98.88 | 91.45 | 89.26 | 98.32 | 97.17 | 91.52 | 92.24 | 94.13 | 92.73 | 93.45 | 89.12 | 95.29 | 93.41 | 90.67 | 96.60 | 92.48 | 96.60 | 95.40 | 95.81 | 95.94 | |
| 8 | 83.59 | 82.70 | 82.32 | 80.92 | 82.48 | 81.59 | 93.78 | 96.60 | 90.86 | 88.17 | 97.17 | 96.00 | 90.94 | 91.66 | 93.56 | 92.57 | 92.88 | 88.24 | 93.51 | 91.60 | 89.38 | 95.43 | 92.10 | 95.42 | 94.20 | 94.58 | 94.74 | |
| 9 | 82.91 | 82.65 | 81.58 | 81.10 | 81.96 | 81.54 | 95.82 | 92.65 | 92.71 | 90.07 | 99.44 | 98.31 | 92.77 | 93.47 | 95.33 | 93.98 | 94.67 | 89.90 | 96.49 | 94.64 | 90.17 | 96.60 | 91.72 | 97.75 | 96.57 | 97.01 | 97.12 | |
| 10 | 96.20 | 93.40 | 97.96 | 92.86 | 92.46 | 92.07 | 81.55 | 82.98 | 82.24 | 96.52 | 92.77 | 92.73 | 94.67 | 94.04 | 95.89 | 96.44 | 96.51 | 85.79 | 92.89 | 90.97 | 87.23 | 90.62 | 87.51 | 92.07 | 91.56 | 92.24 | 92.18 | |
| 11 | 93.10 | 91.13 | 93.27 | 97.60 | 90.33 | 90.93 | 80.64 | 81.40 | 80.87 | 93.62 | 90.17 | 90.11 | 92.15 | 92.74 | 93.40 | 93.95 | 94.06 | 83.78 | 90.32 | 88.31 | 84.83 | 87.41 | 85.38 | 89.42 | 88.92 | 89.63 | 89.56 | |
| 12 | 83.55 | 85.73 | 83.82 | 82.45 | 84.20 | 83.79 | 94.53 | 92.85 | 94.36 | 85.12 | 83.37 | 98.88 | 92.84 | 93.53 | 95.37 | 94.03 | 94.72 | 89.81 | 96.52 | 94.69 | 90.07 | 97.17 | 92.33 | 98.31 | 97.15 | 97.61 | 97.70 | |
| 13 | 83.75 | 85.06 | 83.80 | 83.57 | 84.40 | 84.66 | 92.09 | 91.13 | 93.05 | 84.66 | 84.25 | 93.60 | 92.80 | 93.50 | 95.35 | 94.00 | 94.70 | 88.48 | 95.31 | 93.44 | 88.71 | 96.00 | 91.06 | 97.16 | 95.97 | 96.39 | 96.51 | |
| 14 | 89.11 | 87.57 | 88.86 | 88.90 | 87.58 | 87.57 | 80.55 | 81.09 | 81.50 | 90.48 | 89.32 | 82.15 | 82.82 | 92.83 | 94.71 | 96.54 | 94.63 | 84.22 | 92.96 | 91.63 | 85.55 | 90.22 | 88.21 | 92.22 | 92.28 | 91.35 | 92.32 | |
| 15 | 90.56 | 90.31 | 91.72 | 90.93 | 89.90 | 91.11 | 81.36 | 81.41 | 81.59 | 92.07 | 91.32 | 83.15 | 82.91 | 88.88 | 97.13 | 94.03 | 95.96 | 85.52 | 93.54 | 91.64 | 85.97 | 90.96 | 87.69 | 91.67 | 90.36 | 93.30 | 90.88 | |
| 16 | 91.52 | 92.27 | 92.86 | 92.11 | 91.88 | 93.04 | 82.65 | 82.70 | 82.42 | 92.85 | 92.11 | 84.46 | 83.32 | 89.88 | 96.55 | 95.89 | 98.29 | 87.70 | 95.38 | 93.54 | 88.88 | 92.89 | 90.38 | 94.19 | 92.97 | 94.84 | 93.47 | |
| 17 | 88.93 | 87.62 | 88.69 | 88.31 | 87.62 | 87.40 | 82.47 | 82.53 | 83.16 | 90.11 | 88.73 | 83.57 | 83.32 | 96.00 | 88.08 | 89.29 | 95.83 | 85.11 | 94.13 | 92.24 | 86.45 | 91.44 | 88.16 | 93.41 | 92.22 | 92.57 | 93.50 | |
| 18 | 90.16 | 89.91 | 91.33 | 91.91 | 89.29 | 89.70 | 80.42 | 81.42 | 81.13 | 91.68 | 90.73 | 83.16 | 82.23 | 87.83 | 93.04 | 93.61 | 87.45 | 87.52 | 94.73 | 92.86 | 88.06 | 92.19 | 89.64 | 94.11 | 92.85 | 94.18 | 93.37 | |
| 19 | 73.75 | 75.70 | 74.26 | 74.00 | 76.20 | 75.94 | 78.99 | 78.08 | 79.46 | 75.25 | 73.75 | 80.12 | 79.21 | 72.87 | 74.27 | 75.24 | 73.97 | 74.03 | 86.93 | 85.49 | 86.53 | 89.15 | 87.52 | 89.01 | 87.66 | 89.11 | 88.16 | |
| 20 | 85.52 | 86.53 | 86.34 | 85.03 | 85.67 | 86.10 | 84.01 | 84.50 | 83.56 | 86.54 | 84.60 | 86.59 | 84.22 | 84.30 | 87.62 | 88.01 | 85.89 | 85.49 | 76.64 | 98.29 | 87.40 | 94.13 | 90.22 | 94.72 | 94.67 | 95.16 | 94.59 | |
| 21 | 84.88 | 86.55 | 85.71 | 84.17 | 85.26 | 85.25 | 83.58 | 84.96 | 83.13 | 85.69 | 83.72 | 86.18 | 84.24 | 82.96 | 85.94 | 86.34 | 84.59 | 85.08 | 76.91 | 97.43 | 85.89 | 92.24 | 88.81 | 92.85 | 92.78 | 93.26 | 92.68 | |
| 22 | 75.28 | 75.82 | 75.60 | 75.85 | 75.82 | 76.76 | 81.32 | 79.02 | 79.68 | 75.57 | 75.09 | 78.67 | 78.22 | 76.02 | 75.10 | 77.58 | 76.59 | 75.31 | 73.45 | 77.33 | 75.85 | 89.08 | 86.35 | 88.99 | 87.54 | 88.51 | 88.13 | |
| 23 | 78.14 | 81.27 | 78.93 | 79.16 | 81.51 | 79.85 | 84.98 | 83.26 | 83.22 | 80.34 | 79.90 | 84.29 | 84.32 | 79.81 | 80.56 | 81.22 | 80.99 | 79.62 | 76.30 | 82.60 | 81.48 | 82.79 | 90.46 | 95.43 | 94.20 | 94.58 | 94.74 | |
| 24 | 75.23 | 77.14 | 75.23 | 75.22 | 77.14 | 79.08 | 78.62 | 77.21 | 79.05 | 76.70 | 74.48 | 79.81 | 79.37 | 78.11 | 75.24 | 77.66 | 78.30 | 76.70 | 72.71 | 79.05 | 77.88 | 75.09 | 77.06 | 91.75 | 90.45 | 90.29 | 91.62 | |
| 25 | 81.60 | 81.34 | 80.01 | 80.48 | 81.34 | 81.80 | 89.17 | 90.01 | 90.38 | 80.93 | 80.72 | 90.16 | 89.36 | 81.76 | 80.50 | 81.39 | 82.73 | 80.51 | 77.52 | 83.36 | 82.70 | 78.94 | 83.24 | 78.16 | 97.75 | 97.69 | 99.43 | |
| 26 | 81.11 | 80.61 | 80.46 | 80.92 | 80.13 | 80.84 | 88.54 | 88.98 | 88.92 | 80.90 | 80.22 | 89.94 | 89.35 | 81.32 | 79.76 | 80.66 | 82.30 | 79.77 | 77.50 | 83.16 | 82.27 | 79.88 | 83.89 | 78.62 | 93.79 | 96.57 | 97.69 | |
| 27 | 80.96 | 81.16 | 80.06 | 81.23 | 80.69 | 80.92 | 88.79 | 90.04 | 90.03 | 80.98 | 80.77 | 90.58 | 89.42 | 79.50 | 80.31 | 81.21 | 80.98 | 81.21 | 78.49 | 84.08 | 83.65 | 77.28 | 82.19 | 77.48 | 95.66 | 92.30 | 97.05 | |
| 28 | 81.75 | 81.95 | 81.33 | 80.86 | 81.95 | 82.17 | 88.82 | 88.03 | 89.46 | 81.99 | 80.87 | 88.17 | 87.18 | 80.04 | 78.98 | 80.84 | 81.03 | 79.24 | 78.10 | 82.13 | 81.70 | 78.99 | 83.06 | 77.04 | 93.25 | 90.36 | 91.95 | |
| Nucleotide similarity (%) | ||||||||||||||||||||||||||||
- Note: (1): O-VN-CTU-VL01; (2): O-VN-CTU-VL02; (3): O-VN-CTU-TV01; (4): O-VN-CTU-TV02; (5): O-VN-CTU-TV03; (6): O-VN-CTU-TV04; (7): O-VN-CTU-BT01; (8): O-VN-CTU-BT02; (9): O-VN-CTU-BT03; (10): O-VN-CTU-BT04; (11): O-VN-CTU-BT05; (12): HQ116173.1_O/CAM/5/2006; (13): HQ116274.1_O/VIT/7/2004; (14): HQ116256.1_O/TAI/1/2009; (15): HQ116278-O/VIT/4/2005; (16): JQ070319.1_MYA/3/2010; (17): JQ070325.1_MOG/CO3/2010; (18): KC438373.1_SKR/12/2010; (19): AF154271.1_Tau-YuanTW97, (20): AJ294906.1_O/CAM/11/94; (21): AJ294929.1_O/VIT/2/97; (22): AJ303481.1_O_ALG/1/99; (23): AJ303511.1_O_KEN/83/79; (24): K01201.1_O1Campos/_Brazil/71; (25): KC506471.1_O/IND129/2010; (26): KC506543.1_O/IND62/2011; (27): KY399464.1_O/VIT/9595/2015; (28): MT936117.1_IC491/2018-Ind2001e.
Those results illustrate the circulation of FMDVs lineage Mya-98 (O/SEA) and lineage Pan Asia (O/ME-SA) in the Mekong Delta Provinces, Vietnam.
Figure 3 shows that a total of 15/210 (7.14%) and 30/210 (14.29%) amino acid substitutions were recorded between the predicted VP1 protein of the isolated sequences and the reference sequences within the Pan Asia and Mya-98 lineages, respectively.

Comparison of amino acid sequences of the main antigenic site in the VP1 protein (amino acid positions 133–158) showed that there were some substitutions occurred at Positions 133, 135, 138, 139, 141, and 142 between isolated and FMDV reference strains in lineage Pan Asia and at Positions 137, 138, 140, 143, 144, 152, and 153 in lineage Mya-98. Compared to reference strains in other countries, a substitution mutation was detected at Position 139 in the isolated FMDVs. Remarkably, there was a substitution mutation at Position M144V in FMDVs O-VN-CTU-VL1 (PP897837) and FMDV O-VN-CTU-TV1 (PP897840). The result also showed that amino acid Positions 145–147 (RGD) of isolated and reference FMDV strains were completely conserved.
Furthermore, the amino acid differences of the isolated FMDVs with FMDV vaccine strain O1/Campos/Brazil (K01201.1) were compared in this study. Compared with the vaccine strain, the substitution at Positions 133, 137, 138, 139, 140, 141, 142, and 155 were detected in the isolated strains in lineage Pan Asia and at Positions 135, 137–144, 152, and 153 in lineage Mya-98.
3.3. Recombination Analysis
The negative recombinant events in VP1 sequences of isolated FMDVs were characterized by using a standard similarity plot (Simplot) and RDP.
4. Discussion
Among infectious diseases in cattle, FMD is one of the most important diseases of livestock and is endemic in several countries broadly. The FMD also causes economic losses to the livestock population and reduces food security and economic development. FMD has been prevalent in Vietnam since 1898 with occurrences of sporadic outbreaks from time to time. Several studies on FMD have been published in Northern Vietnam; however, no studies have determined the evolution of FMDVs through a complete analysis in the Mekong Delta Provinces. The interpretation of the phylogenetic relationships among the FMDV strains contributes to proper control measures against the disease. Therefore, the present study considered it necessary to fill the existing knowledge gap of characterization, evolution, and origins of FMDV strains. The study results would be critical for developing and deploying effective vaccines in the future.
The VP1 sequences are usually used to identify the molecular characterization of FMDV [22]. Besides, the VP1 capsid protein plays an important role in the investigating relationship between different FMDV strains because of its significance for protective immunity, serotype specificity, and attachment and entry of FMDV. In the present study, the VP1 nucleotide and amino acid sequences of the FMDV Serotype O from isolated strains in the Mekong Delta Provinces in Vietnam and published reference strains in the north and northern central regions of Vietnam and other Asian countries (China, Laos, Cambodia, Myanmar, Bangladesh, Mongolia, South Korea, and Thailand) (Table 1) were used for the identification of the phylogeny and genetic relationships between isolated FMDVs and reference strains.
In this study, phylogenetic analysis and PSCA showed that there was the circulation of FMDV lineage Mya-98 (O/SEA) (O-VN-CTU-VL01 [PP897837], O-VN-CTU-VL02 [PP897838], O-VN-CTU-TV01 [PP897840], O-VN-CTU-TV02 [PP897841], O-VN-CTU-TV03 [PP897842], O-VN-CTU-TV04 [PP897844], O-VN-CTU-BT04 [PP897847], and O-VN-CTU-BT05 [PP897847]); and lineage Pan Asia (O/ME-SA) (O-VN-CTU-BT01 [PP897844], O-VN-CTU-BT02 [PP897844], and O-VN-CTU-BT03 [PP897844]) in the Mekong Delta Provinces. This result is consistent with a previously published study in [23]; Pan Asia (O/ME-SA) and Mya-98 (O/SEA) were different lineages of Serotype O which have been circulating in Vietnam recently.
Topotype SEA was the most frequently detected in Type O and had two lineages, named Mya-98 and Cam-94 [24]. The isolated FMDV strain, lineage Mya-98 (O/SEA), shared a close relationship with those strains from the north and northern central regions of Vietnam and Asia countries consisting of China, South Korea, Myanmar, Thailand, and Mongolia. These findings are partially consistent with previous studies conducted by the OIE, which assessed the livestock movement in Vietnam and neighboring countries [25]. In addition, previous reports showed that Mya-98 (O/SEA) was normally restricted to all six mainland countries of the SEA region and has caused outbreaks in Myanmar (1998), Laos (1998), Cambodia (1998), Thailand (1999), Malaysia (2001), and Vietnam (2002) [22]. Besides, Mya-98 (O/SEA) has caused outbreaks in eastern and Central Asia, specifically Hong Kong (2010), South Korea (2010–2011), North Korea (2010), Japan (2010), Mongolia (2010), Russia (2010), and Taiwan (2012) [26–28].
Lineage Pan Asia (O/ME-SA) was first described in the 1980s in the Indian subcontinent and subsequently disseminated broadly throughout southern Asia, the Middle East, and East and Southeast Asia, and some incursions in South Africa and European countries [8]. Moreover, the Pan Asia lineage had previously been introduced into SEA specifically in Malaysia in 2003–2005 and became widespread along with the local Mya98 (O/SEA). Besides, Pan Asia was found during this period in Cambodia, Vietnam, Lao, China, and Thailand [29].
Our findings indicate that Mya-98 (O/SEA) and Pan Asia (O/ME-SA) are the predominant lineages or topotypes of FMDV Serotype O circulating in the Mekong Delta Provinces (southern Vietnam).
The analysis of amino acid sequences of VP1 indicates several substitution mutations detected in isolated FMDVs, especially, mutations at the antigenic site (amino acid position 133–158). According to [30], the peptide 136–151 plays an important role in neutralizing FMDV, and particularly amino acid 139 is a decisive factor in the neutralizing properties of the main antigen region in VP1. In this study, a substitution mutation at Position M144V was recorded in FMDVs O-VN-CTU-VL1 (PP897837) and FMDV O-VN-CTU-TV1 (PP897840). A previous study of [31] revealed that the amino acids 140–160 are the first neutralizing antigen site in VP1 of FMDV, in which the amino acids 144, 148, and 154 play a decisive role in the neutralizing properties of this antigenic site of FMDV. Furthermore, a change in any of the three amino acids 144, 154, or 208 can lead to a decrease in the ability to neutralize the VP1 antigen of FMDV [32]. Therefore, further study in FMDVs O-VN-CTU-VL1 (PP897837) and FMDV O-VN-CTU-TV1 (PP897840) should be performed. In the present study, 3 amino acid RGD at Positions 145–147 were detected in all FMDVs. According to [33], the amino acid sequence involved in the attachment of FMDV Serotype O to cells is located in regions 141–160 and 200–213, in which amino acids 145–147, 206, and 207 play a decisive role.
In Vietnam, cattle are compulsorily vaccinated against FMDV Serotype O twice annually with varying vaccine formulations, based on current knowledge of circulating strains. However, sporadic FMD outbreaks have been reported in southern Vietnam. This suggests there may be a vaccination failure. Possible causes of vaccination failure include the presence of immunosuppressive disorders, improper storage and transport of the vaccine, and immune evasion due to the presence of different vaccine variants as previously reviewed [34]. Besides, changes in the antigenic properties should be considered. According to [35–37], antigenic properties of FMDV strains are associated with specific amino acid motifs in the VP1 protein: 43–45, 83, 96, 141–160 (G-H loop), 169–173, and 200–211 (C-terminus). In this study, substitutions at Positions 133, 137–142, and 155 were detected in the isolated strains in lineage Pan Asia (O/ME-SA) and at Positions 135, 137–144, 152, and 153 in lineage Mya-98 (O/SEA). Hence, the possibility that the current vaccine strains may offer reduced protection against the recent circulating strains. Cross-neutralization studies should be assessed in further studies to evaluate the effectiveness of current vaccines in the prevention of FMD in southern Vietnam.
5. Conclusions
Molecular characterization of VP1 sequences of circulating FMDVs in the Mekong Delta Provinces, Vietnam, has provided valuable information on the circulation and evolution of FMDVs. Genetic analysis of nucleotide and amino acid sequences of VP1 illustrated substitution mutation at the antigenic region contributing to the emergence of new FMDVs in lineage Mya-98 (O/SEA) and Pan Asia (O/ME-SA) circulating in the Mekong Delta Provinces, Vietnam. Moreover, further studies about the pathogenicity of FMDVs O-VN-CTU-VL1 (PP897837) and FMDV O-VN-CTU-TV1 (PP897840) should be performed for insight knowledge about these isolates contributing to vaccine development for controlling the FMD.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This study was supported by the Can Tho University grant (B2022-TCT-13) from the Ministry of Education and Training in Vietnam.
Acknowledgments
This study was supported by the Can Tho University grant (B2022-TCT-13) from the Ministry of Education and Training in Vietnam.
Open Research
Data Availability Statement
All data generated or analyzed during this study are included in this published article.




