Aloe vera Phytochemicals as Potential Antibacterial Agents Against Multidrug-Resistant Pseudomonas aeruginosa
Abstract
Multidrug-resistant (MDR) Pseudomonas aeruginosa poses a global challenge due to its high virulence and resistance mechanisms, which lead to persistent infections. This study explored the efficacy of active compounds of Aloe vera as alternative treatments against MDR P. aeruginosa isolates. A total of 283 P. aeruginosa isolates were obtained from sputum and throat samples. The antibiotic resistance of these isolates was evaluated against several antibiotic classes. The minimum inhibitory concentration (MIC) of A. vera–derived phytochemicals was individually assessed; lupeol was found to be a potent phytochemical with the lowest MIC (125 μg/mL). The physicochemical properties and toxicity of the phytochemicals were further evaluated using SwissADME, StopTox, and Protox 3.0. Genetic analysis identified mutations in the AmpC protein as a key factor of antibiotic resistance; consequently, molecular docking studies examined interactions between A. vera phytochemicals and AmpC. Most P. aeruginosa isolates exhibited pronounced antibiotic resistance, while the phytochemicals demonstrated favorable pharmacological properties and strong binding interactions with key amino acids in AmpC. Of these phytochemicals, aloe-emodin showed the most promising antibacterial activity. These findings underscore the potential of A. vera–derived phytochemicals for clinical application as alternative treatments for combating drug-resistant bacterial infections.
1. Introduction
The increasing prevalence of multidrug-resistant (MDR) bacteria presents a significant global challenge to public health and complicates the treatment of community-acquired and healthcare-associated infections [1–3]. Gram-negative bacteria mainly exhibit intrinsic resistance mechanisms, thus posing substantial treatment challenges [4]. Pseudomonas aeruginosa (P. aeruginosa) is a ubiquitous opportunistic pathogen and a prominent example of a MDR Gram-negative bacterium that seriously threatens immunocompromised individuals due to its role in various severe infections [5]. It frequently causes infections, including respiratory tract, gastrointestinal, systemic, and wound infections, in healthcare settings. Notably, P. aeruginosa can shift from a commensal state to a pathogenic form that causes persistent and challenging infections in susceptible populations [6]. It is a primary cause of hospital-acquired pneumonia, with mortality rates of ventilated patients reaching up to 50%. It presents significant risks to individuals with cystic fibrosis [7] or advanced chronic obstructive pulmonary disease, as well as to those in intensive care or with burn injuries [8].
Infections contracted in healthcare settings, including bacteremia, urinary tract infections, pneumonia, and surgical site infections, are often attributed to P. aeruginosa and are estimated to account for 7.1%–7.3% of healthcare-associated illnesses [9, 10]. Epidemiological studies indicate a growing trend in antimicrobial resistance (AMR), with the emergence of MDR bacteria in recent years [11, 12]. The extensive use of antibiotics has significantly advanced healthcare and saved countless lives; however, it has also hastened the rise of MDR bacteria, which present a grave challenge to healthcare systems worldwide [13].
P. aeruginosa employs various virulence factors and rapidly develops antibiotic resistance, thereby enhancing its ability to invade host tissues and increasing infection risk. To enhance its drug resistance and evade the host’s immune responses, it produces an extracellular polysaccharide matrix for protection, which prolongs infections and aids bacterial adherence to surfaces, thus enabling colonization [14]. It is a major MDR pathogen and has been designated a high-priority pathogen by the World Health Organization (WHO) due to its substantial antibiotic resistance and serious threat to public health [15].
The ability of P. aeruginosa to withstand common antibiotics, driven by both phenotypic and genotypic traits, poses substantial financial burdens and jeopardizes patient survival [16]. It possesses a wide range of metabolic abilities, including the ability to form biofilms on various living and nonliving surfaces, a trait that enables it to evade immune responses and significantly complicates treatment. This adaptability promotes persistent infections and drives recurrent AMR, further worsening treatment outcomes and posing a considerable challenge in clinical settings [17]. P. aeruginosa uses protective strategies, such as decreasing outer membrane permeability, employing efflux systems, and generating enzymes that neutralize antibiotics. Resistance may develop through genetic mutations or the acquisition of genes via horizontal gene transfer [18, 19].
The ineffectiveness of traditional antibiotics against MDR pathogens underscores the need for new therapeutic options, particularly bioactive compounds from natural sources. Medicinal plants have been found to show antibacterial activity in vitro and in vivo [20]. The WHO estimates that 80% of individuals in developing nations depend on traditional remedies and plant-based extracts for treating infections [21]. Medicinal plants with antimicrobial and anti-inflammatory properties have become prominent as alternative treatments for infectious diseases in recent years. Aloe vera (L.) Burm.f. (syn. A. barbadensis Mill.; Asphodelaceae) is particularly notable for its longstanding use and therapeutic properties, making it a valuable candidate for treating human infections [22]. It is known for its biological benefits, including immune-modulating, antioxidant, anti-inflammatory, and antibacterial properties, with over 75 active compounds identified in its inner gel [23–25].
This study investigated the antibiotic resistance of P. aeruginosa isolates while evaluating the potential of A. vera and its active compounds as a possible remedy for drug-resistant isolates. The dual focus addressed the issue of resistant bacteria and explored potential solutions provided by A. vera. Although the pharmacological properties of A. vera gel are well documented, its potential antimicrobial applications remained unexplored, which warrants further investigation. This study included molecular detection and determination of the AMR spectrum of highly resistant P. aeruginosa isolated from respiratory infections. In addition, it applied computational approaches, including ADME (absorption, distribution, metabolism, and excretion) prediction and molecular docking, to investigate the inhibitory potential and pharmacokinetic properties of A. vera’s bioactive compounds.
2. Materials and Methods
2.1. Study Design and Specimen Collection
This cross-sectional study included 660 sputum and throat swab samples collected from primary care facilities and tertiary hospitals in Pakistan. It ensured that the patients had not undergone any previous antibiotic treatment. The study received ethical approval from the institutional review committee of the University of Lahore, Pakistan, and adhered to the World Medical Association’s Declaration of Helsinki guidelines [26]. Sterile containers with Amies medium and swabs were employed to transport the samples to the laboratory. Upon arrival at the laboratory, the samples were inoculated onto nutrient agar and incubated overnight at 37°C. Well-isolated single colonies were selected and transferred to agar plates (blood, chocolate, and MacConkey), which were then incubated at 37°C for 24–48 h. The suspected colonies were subsequently subcultured for additional examination and identification.
2.2. Identification of Bacterial Isolates
P. aeruginosa isolates were identified by colony morphology and standard biochemical tests. API 20NE (bioMérieux, Craponne, France) was employed to identify P. aeruginosa isolates [27]. In addition, the isolates underwent molecular characterization to verify the presence of P. aeruginosa. The conventional boiling method was employed to extract DNA from the bacterial samples, which was then subjected to PCR amplification. The OprI gene (249 bp) was amplified using the primers OprI-F (5 ′-ATGAACAACGTTCTGAAATTCTCTGCT-3 ′) and OprI-R (5 ′-CTTGCGGCTGGCTTTTTCCAG-3 ′), while the OprL gene (504 bp) was amplified with the primers OprL-F (5 ′-ATGGAAATGCTGAAATTCGGC-3 ′) and OprL-R (5 ′-CTTCTTCAGCTCGACGCGACG-3 ′) [28–30]. The PCR reactions were performed at a total volume of 25 μL. This mixture consisted of 11 μL of DNase, 8 μL of 2X PCR Master Mix with 1.5 mM MgCl2, 0.5 μL of each primer, and 5 μL of DNA template. The thermal cycling procedure started with a 5-min initial denaturation step at 94°C. This was followed by 30 repetitions of a three-step cycle: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The procedure concluded with a final extension phase lasting 10 min at 72°C. Controls included positive samples (P. aeruginosa DNA) and negative samples (water). The PCR products were separated using 1% agarose gels in TBE buffer (40 mM Tris, 20 mM boric acid, and 1 mM EDTA at pH 8.3) and visualized under UV light [31].
2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility against all the major classes of antibiotics was assessed using the Kirby–Bauer disc diffusion method. A variety of standard antibiotic discs (Oxoid, United Kingdom) were used, including imipenem (10 μg), meropenem (10 μg), piperacillin/tazobactam (100/10 μg), ticarcillin/clavulanic acid (75/10 μg), cefepime (30 μg), cefoperazone (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (5 μg), amikacin (30 μg), gentamicin (10 μg), tobramycin (10 μg), tigecycline (4 μg), ciprofloxacin (5 μg), and levofloxacin (5 μg). The ATCC 27853 strain (GenBank accession CP015117) of P. aeruginosa was employed for quality control (QC) purposes. The findings were analyzed according to the guidelines established by the Clinical and Laboratory Standards Institute (CLSI) [32]. The isolates that showed resistance against at least three antibiotic classes were declared MDR, and those against at least five classes were declared extensively drug-resistant (XDR). The isolates that resisted all the tested classes of antibiotics were considered pan-drug-resistant.
2.4. Determining the Antibacterial Efficacy of Aloe vera–Derived Phytochemicals Using Agar Well Diffusion Assay
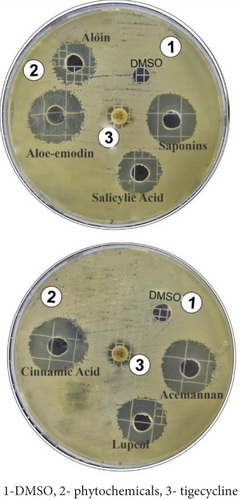
Based on an extensive literature review, the antibacterial activity of A. vera phytochemicals, including aloin, aloe-emodin, acemannan, saponins, lupeol, salicylic acid, and cinnamic acid, was evaluated against P. aeruginosa using the agar well diffusion method. P. aeruginosa was cultured in nutrient broth at 37°C, and the bacterial culture was used when it reached the log phase of growth. Wells 6 mm in diameter were created using a sterile borer in Mueller–Hinton agar plates. A 100-μL amount of bacterial suspension was uniformly spread using a sterile cotton swab in a circular motion across the agar surface to form a bacterial lawn. Phytochemicals, such as aloin, aloe-emodin, acemannan, saponins, lupeol, salicylic acid, and cinnamic acid, were dissolved in DMSO at a concentration of 2 μg/mL, and 100 μL of each phytochemical solution was placed into the designated wells. The plates were incubated at 37°C for 24 h. All experiments were conducted in triplicate, and the inhibition zones were measured according to CLSI standards [32].
2.5. Evaluation of the Minimum Inhibitory Concentration (MIC) of Aloe vera Phytochemicals
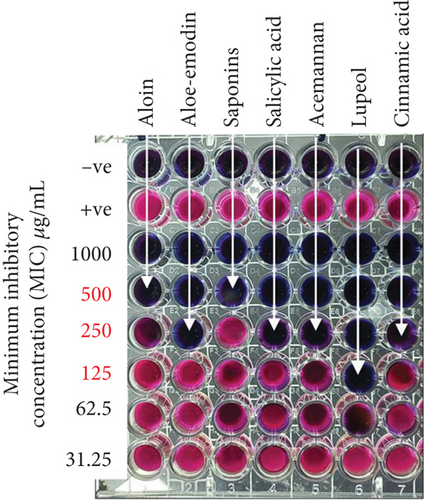
The antibacterial activity of A. vera phytochemicals against MDR P. aeruginosa was evaluated using a MIC assay in a 96-well microtiter plate. Phytochemicals such as aloin, aloe-emodin, acemannan, saponins, lupeol, salicylic acid, and cinnamic acid were dissolved in DMSO with a 1000 μg/mL stock. Two-fold serial dilutions, ranging from 1000 to 31.25 μg/mL, were made. The MDR P. aeruginosa was grown in Mueller–Hinton broth (MHB) at 37°C until it reached the mid-logarithmic growth phase. The bacterial suspension was standardized to a 0.5 McFarland standard (1.5 × 108 CFU/mL). In the microtiter plate, 100 μL of MHB was added to each well, followed by 100 μL of each phytochemical, beginning with the highest concentration and proceeding through two-fold serial dilutions. Then, 50 μL of the bacterial suspension was added to the wells containing the phytochemicals. The positive control wells contained only the bacterial suspension without any phytochemicals, while the negative controls had only MHB. The plate was incubated for 24 h at 37°C. After incubation, 20 μL of resazurin dye was introduced into each well and allowed to react for 2–4 h. A blue color indicated the absence of bacterial growth, whereas a pink color suggested active growth. The MIC was determined as the lowest phytochemical concentration that prevented visible bacterial growth, indicated by the absence of a color change. All experiments were conducted in triplicate.
2.6. Statistical Analysis
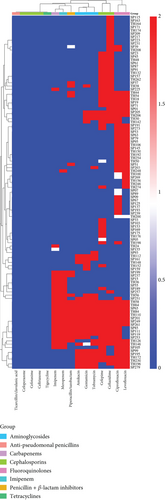
Statistical analyses were performed using SPSS version 25.0. Frequencies were determined for qualitative variables, while the mean ± standard deviation (SD) was calculated for quantitative variables. The chi-square test was employed to evaluate associations between categorical variables, and one-way analysis of variance (ANOVA) was conducted to compare the mean inhibition zones between the different phytochemicals and tigecycline. Both have a significant level determined with a p value ≤ 0.05. A heat map illustrating antibiotic resistance profiles of P. aeruginosa isolates was generated in R (4.4.2) using the heat map package.
2.7. Evaluation of Drug Profile Through ADME and Toxicity Analyses
Pharmacokinetic and absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties were assessed using the pkCSM web server (http://structure.bioc.cam.ac.uk/pkcsm) and SwissADME server (http://www.swissadme.ch; accessed on September 4, 2024). A molecular fingerprinting approach was applied to evaluate drug-likeness, based on Lipinski’s rule of five (LRF). Toxicological assessments were conducted using the ProTox-3.0 (ProTox-3.0—Prediction of Toxicity of chemicals (charite.de)) servers and StopTox (https://stoptox.mml.unc.edu/), which evaluated various toxicity endpoints using QSAR models and machine learning methods [33].
2.8. Retrieval of Ligands and Receptors
The crystal structure of wild-type AmpC protein (PDB ID: 7FF0) was obtained from the Protein Data Bank (RCSB PDB; https://www.rcsb.org; accessed on September 4, 2024) in PDB format. The structural representations of these therapeutically significant compounds were obtained from the PubChem database. ChemDraw Professional (Version 21.0) was employed to construct 2D and 3D structures of these compounds, while Discovery Studio Visualizer (v21.1.0.20298) was used for their visualization [34, 35].
2.9. Protein Preparation
The protein structure was imported from the PDB, and any structural defects in the residues were corrected. The Schrödinger suite (Maestro Wizard version 21.5) was used to process the proteins. Initially, preprocessing was performed by filling the gap between loops and side chains by applying prime job, adding hydrogen atoms, and creating disulfide bonds at a constant pH of 7.0 ± 2. Preprocessing was followed by H-bond optimization using PROPKA at an optimized pH of 7.0; subsequently, water molecules in a radius of 3.0 Å were removed. Finally, the structure was minimized using the OPLS3e force field. The scaling factor for the van der Waals (vdW) radius was adjusted to 1.0, and grid files outlining the receptor binding sites were created [36].
2.10. Ligand Preparation
The reference compound and ligands were generated using the LigPrep module of Schrödinger. Low-energy three-dimensional conformations were created with suitable chiralities and refined using the OPLS3e force field at a physiological pH of 7.2 ± 0.2. A detailed protocol by Rauf et al. was followed [37].
2.11. Protein–Ligand Docking
Protein–ligand interactions were analyzed using Maestro because of its enhanced precision. The binding site’s orientation and dimensions for ligand docking were determined by constructing a receptor grid. The Schrödinger suite was used to generate the scoring grid based on the AmpC crystal structure (Maestro Wizard version 21.5). Molecular docking was performed using Glide in Schrödinger Maestro 21.5 and employed both standard precision (SP) and extra precision (XP) approaches, along with flexible ligand sampling. The cutoff for partial charges on ligand atoms was set to 0.15, while the scaling factor for vdW radii was set to 0.80. Additionally, root-mean-square deviation (RMSD) value was also computed to validate the docking accuracy [36].
2.12. Visualization of Protein–Ligand Complexes
AmpC protein structures were visualized using UniProt and AlphaFold. Discovery Studio Visualizer 20.1 was used to visualize protein–ligand complexes and identify interactions. Docked structures were also analyzed using PyMOL (Version 2.4.0).
2.13. Density Functional Theory (DFT) Analysis
DFT computations were used to investigate the physicochemical characteristics of the chosen compounds. The RB3LYP method was employed for both optimization and frequency analysis using the Gaussian 09 W program. The energy levels of the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), as well as the band gap, were determined using the same theoretical framework to derive additional quantum chemical parameters. The energy band gap (ΔEGap = ELUMO − EHOMO) was obtained by calculating the difference between the LUMO and HOMO energy values and visualized on Gauss View 5.0.8 software. Furthermore, the DFT method was used to evaluate the molecular electrostatic potential of the compounds by using a checkpoint (.chk) file [38]. A detailed protocol by Rauf et al. was followed [39].
3. Results
3.1. Specimens and Isolates
This study included 660 patients from diverse ethnic backgrounds. Of these, 387 (58.6%) were male, and 273 (41.4%) were female. The incidence of P. aeruginosa infection was significantly higher in males (54%) compared to females (46%) (p = 0.05). The participants’ ages ranged from 1 to 95 years, with a mean age of 50.31 ± 21.113 years. The prevalence of P. aeruginosa varied by age group, with the highest prevalence in Age Group IV (61–80 years) at 31.41% (n = 89), followed by Age Groups II (21–40 years) and III (41–60 years) at 26.9% (n = 76) and 27.5% (n = 77), respectively (Table 1). The lowest prevalence was observed in Age Group I (1–20 years) at 8.3% (n = 25) and Age Group V (81–100 years) at 5.65% (n = 16). The differences in prevalence between age groups were statistically significant (p = 0.02). Regional distribution showed 18 cases (2.8%) from Azad Jammu and Kashmir, 37 cases (5.6%) from Khyber Pakhtunkhwa, 573 cases (86.8%) from Punjab, and 32 cases (4.8%) from Sindh. The highest infection rate was in the Pakhtoon ethnic group at 30% (n = 238), followed by the Sindhi (28%), Punjabi (22%), and Kashmiri (20%) groups.
| Variable | Total cohort n (%) (n = 660) | P. aeruginosa–infected cases, n (%) (n = 283) | pvalue |
|---|---|---|---|
| Age group (years) | |||
| > 20 | 71 (10.7) | 23 (8.1) | 0.257 |
| 21–40 | 147 (22.1) | 71 (25.1) | |
| 41–60 | 204 (30.7) | 85 (30.0) | |
| 61–80 | 207 (31.2) | 92 (32.5) | |
| 81–100 | 31 (4.7) | 12 (4.2) | |
| Gender | |||
| Male | 387 (58.3) | 147 (51.9) | 0.007 |
| Female | 273 (41.1) | 136 (48.1) | |
| Province | |||
| Punjab | 578 (87) | 248 (87.6) | 0.259 |
| Sindh | 32 (4.8) | 17 (6.0) | |
| Khyber Pakhtunkhwa | 36 (5.4) | 15 (5.3) | |
| Azad Jammu and Kashmir | 14 (2.1) | 3 (1.1) | |
| Antibiotic resistance | |||
| Imipenem | 92 (13.9) | 26 (9.1) | 0.005 |
| Meropenem | 92 (13.9) | 25 (8.8) | 0.001 |
| Piperacillin/tazobactam | 104 (15.7) | 21 (7.4) | < 0.001 |
| Ticarcillin/clavulanic acid | 0 (0) | 0 (0) | — |
| Cefepime | 77 (11.6) | 52 (18.3) | < 0.001 |
| Cefoperazone | 0 (0) | 0 (0) | — |
| Cefotaxime | 104 (15.7) | 0 (0) | < 0.001 |
| Vancomycin | 0 (0) | 0 (0) | — |
| Ceftazidime | 98 (14.8) | 51 (18.0) | 0.078 |
| Ceftriaxone | 105 (15.9) | 0 (0) | < 0.001 |
| Amikacin | 72 (10.9) | 23 (8.1) | 0.061 |
| Gentamicin | 122 (18.4) | 30 (10.6) | < 0.001 |
| Tobramycin | 102 (15.4) | 28 (9.8) | 0.001 |
| Ciprofloxacin | 261 (39.5) | 56 (19.7) | < 0.001 |
| Levofloxacin | 163 (24.6) | 45 (15.9) | < 0.001 |
| Tigecycline | 8 (1.2) | 0 (0) | 0.014 |
| Coresistance | |||
| Multidrug-resistant | 447 (67.3) | 177 (62.5) | < 0.001 |
| Extensively drug-resistant | 29 (4.4) | 8 (2.8) | |
| Pan-drug-resistant | 15 (2.3) | 0 (0) |
3.2. Microbial Spectrum Associated With Respiratory Origins
The microbial spectrum of respiratory infections revealed the presence of 24 microbial taxa. P. aeruginosa was found to be the most prevalent, followed by Klebsiella pneumoniae and Acinetobacter baumannii. The least prevalent microbial species included Acinetobacter lwoffii, Aspergillus flavus, Aspergillus fumigatus, Citrobacter koseri, Citrobacter spp., and Enterobacter aerogenes (Table S1).
3.3. Antibiotic Susceptibility Profile of P. aeruginosa
The patterns of resistance of P. aeruginosa to all the major antibiotic classes were evaluated. All the isolates were found to be sensitive to ticarcillin/clavulanic acid, cefoperazone, cefotaxime, ceftriaxone, and tigecycline. Ciprofloxacin proved to be the most effective antibiotic, followed by cefepime, ceftazidime, gentamicin, tobramycin, imipenem, meropenem, and piperacillin/tazobactam. The antibiotic resistance patterns of the bacterial isolates were observed to be influenced by the gender, age, and provincial distribution of the patients. In general, the isolates recovered from male patients, from patients in the 41–60-year age group, and from the Punjabi ethnic group were more resistant (Table 2).
| Age-wise distribution of resistant isolates (years), n (%) | Gender-wise distribution of resistant isolates, n (%) | Province-wise distribution of resistant isolates, n (%) | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| > 20 | 21–40 | 41–60 | 61–80 | > 80 | p value | Male | Female | p value | Punjab | Sindh | KPK | AJK | p value | ||
| Antibiotic | |||||||||||||||
| Imipenem | 1 (0.4) | 6 (2.1) | 8 (2.8) | 7 (2.5) | 4 (1.4) | 0.18 | 189 (6.4) | 8 (2.8) | 0.109 | 24 (8.5) | 0 (0) | 0 (0) | 2 (0.7) | 0 | 26 (9.2) |
| Meropenem | 1 (0.4) | 6 (2.1) | 8 (2.8) | 6 (2.1) | 4 (1.4) | 0.038 | 17 (6.0) | 8 (2.8) | 0.092 | 23 (8.2) | 0 (0) | 0 (0) | 2 (0.7) | 0.01 | 25 (8.8) |
| Piperacillin/tazobactam | 0 (0) | 5 (1.8) | 7 (2.5) | 7 (2.5) | 2 (0.7) | 0.67 | 12 (4.2) | 9 (3.2) | 0.551 | 21 (7.4) | 0 (0) | 0 (0) | 0 (0) | 0.76 | 21 (7.4) |
| Ticarcillin/clavulanic acid | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Cefepime | 4 (1.4) | 16 (5.7) | 18 (6.4) | 10 (3.5) | 4 (1.4) | 0.181 | 26 (9.2) | 26 (9.2) | 0.951 | 48 (17) | 2 (0.7) | 2 (0.7) | 0 (0) | 0.925 | 52 (18.4) |
| Cefoperazone | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Cefotaxime | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Ceftazidime | 4 (1.4) | 13 (4.6) | 16 (5.7) | 14 (4.9) | 4 (1.4) | 0.655 | 28 (9.9) | 23 (8.1) | 0.64 | 46 (16.3) | 2 (0.7) | 3 (1.1) | 0 (0) | 0.752 | 51 (18) |
| Ceftriaxone | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Amikacin | 1 (0.4) | 7 (2.5) | 10 (3.5) | 3 (1.1) | 2 (0.7) | 0.176 | 13 (4.6) | 10 (3.5) | 0.647 | 21 (7.4) | 0 (0) | 1 (0.4) | 1 (0.4) | 0.248 | 23 (8.1) |
| Gentamicin | 1 (0.4) | 10 (3.5) | 12 (4.2) | 5 (1.8) | 2 (0.7) | 0.334 | 15 (5.3) | 15 (5.3) | 0.564 | 25 (8.9) | 2 (0.7) | 2 (0.7) | 1 (0.4) | 0.923 | 30 (10.6) |
| Tobramycin | 2 (0.7) | 9 (3.2) | 10 (3.5) | 5 (1.8) | 2 (0.7) | 0.454 | 14 (4.9) | 14 (4.9) | 0.828 | 26 (9.2) | 0 (0) | 1 (0.4) | 1 (0.4) | 0.263 | 28 (9.9) |
| Tigecycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Ciprofloxacin | 3 (1.1) | 16 (5.7) | 21 (7.4) | 14 (4.9) | 2 (0.7) | 0.605 | 22 (7.8) | 34 (12) | 0.056 | 47 (16.5) | 3 (1.1) | 5 (1.8) | 1 (0.4) | 0.828 | 56 (19.8) |
| Levofloxacin | 2 (0.7) | 15 (5.3) | 12 (4.2) | 14 (4.9) | 2 (0.7) | 0.77 | 17 (6.0) | 28 (9.9) | 0.077 | 39 (13.8) | 1 (0.4) | 5 (1.8) | 0 (0) | 45 (15.9) | |
| Coresistance | |||||||||||||||
| Non-MDR | 7 (2.5) | 21 (7.4) | 27 (9.5) | 36 (12.7) | 7 (2.5) | 0.226 | 51 (18) | 47 (16.7) | 0.707 | 85 (30) | 8 (2.8) | 4 (1.4) | 1 (0.4) | 0.846 | 98 (34.6) |
| MDR | 15 (5.3) | 50 (17.8) | 53 (18.7) | 54 (19.1) | 5 (1.8) | 93 (32.9) | 84 (29.7) | 156 (55.1) | 9 (3.2) | 10 (3.5) | 2 (0.7) | 177 (62.5) | |||
| XDR | 1 (0.4) | 0 (0) | 5 (1.8) | 2 (0.7) | 0 (0) | 3 (1.1) | 5 (1.8) | 7 (2.5) | 0 (0) | 1 (0.4) | 0 (0) | 8 (2.8) | |||
- Note: KPK, Khyber Pakhtunkhwa; MDR, multidrug-resistant; XDR, extensively drug-resistant.
- Abbreviation: AJK, Azad Jammu and Kashmir.
3.4. Prevalence of Coresistant P. aeruginosa
Of the 283 P. aeruginosa isolates, 177 (63.2%) were MDR and 8 (2.9%) were XDR. The distribution of coresistant isolates varied according to ethnic group, age, and gender of the patients. The highest MDR prevalence was observed in the Punjabi group, followed by the Pakhtoons, Sindhi, and Kashmiris. The prevalence of XDR isolates was highest in the Punjabi group (2.5%) and lowest in the Kashmiri and Sindhi groups (0.0%). Non-MDR isolates were most prevalent in the Punjabi group and least prevalent in the Kashmiri group. However, these differences in prevalence were not statistically significant. Moreover, male patients were observed to harbor more non-MDR and MDR isolates, but an inverse trend was observed for XDR. Furthermore, the prevalence of non-MDR and MDR isolates was observed to increase with increasing age of the patients up to 80 years. However, XDR isolates were more prevalent in the 41–60-year age group (Table 2). In addition, a cluster analysis was performed to assess both the efficacy of each antibiotic in combating P. aeruginosa and the overall sensitivity pattern across all tested antibiotics. The analysis of bacterial isolate clustering based on their AMR patterns revealed considerable diversity. The dendrogram in Figure 1 visualizes the grouping of P. aeruginosa isolates according to their AMR profile. The extent of branching in the dendrogram reflects the variation in resistance, with more diverse branches indicating greater variability between the isolates.
3.5. Antibacterial Efficacy of Aloe vera Phytochemicals Determined Using Agar Well Diffusion Assay
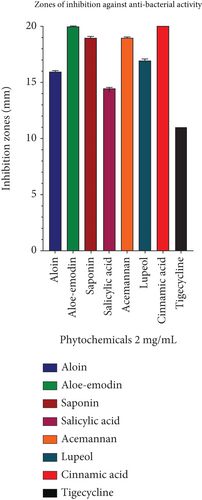
The antibacterial activity of A. vera phytoconstituents against the PDR strain of P. aeruginosa was determined at a concentration of 2 mg/mL. The maximum zone of inhibition of cinnamic acid was 21.10 ± 0.17 mm, while those of acemannan 18.93 ± 0.11 and lupeol were 16.9 ± 0.17 mm each, aloe-emodin was 19.93 ± 0.11 mm, and saponins were 18.93 ± 0.11 mm. The smallest inhibition zones were recorded for aloin and salicylic acid because they inhibited growth only by 15.93 ± 0.11 mm and 14.43 ± 0.11 mm, respectively (Table 3, Figure 2). Tigecycline (15 μg/Disc) is used as a standard antibiotic, showing an 11 ± 00 mm inhibition zone. The results of the ANOVA test (p < 0.0001) ensure that the differences between phytochemicals and tigecycline inhibition are highly significant (Table 4).
| Phytochemicals | Aloin | Aloe-emodin | Saponin | Salicylic acid | Acemannan | Lupeol | Cinnamic acid | Tigecycline |
|---|---|---|---|---|---|---|---|---|
| Inhibition zones | 15.93 ± 0.11 | 19.93 ± 0.11 | 18.93 ± 0.11 | 14.43 ± 0.11 | 18.93 ± 0.11 | 16.9 ± 0.17 | 21.10 ± 0.17 | 11 ± 00 |
| ANOVA summary | Results |
|---|---|
| p value | < 0.0001 |
| p value summary | ∗∗∗∗ |
| R-squared | 0.9989 |
3.6. The MIC of Aloe vera Phytoconstituents
The MICs of A. vera phytochemicals against MDR P. aeruginosa were as follows. Lupeol had the lowest MIC of 125 μg/mL, thus making it the most potent of all the phytochemicals. Aloe-emodin, acemannan, salicylic acid, and cinnamic acid had moderate MICs, with all having values of 250 μg/mL. Steroidal saponins and aloin had the highest MIC of 500 μg/mL, thus indicating that they were the least effective compared to the other phytoconstituents (Figure 3). The antibiotic imipenem/cilastatin 100 μg/mL was used as a standard positive control. The p value is < 0.0001, suggesting statistical significance in the differences between groups. The three asterisks indicate a significant result. There is an R-squared of 0.9979, which means the model explains 99.79% of the variability in the dependent variable. A very high R-squared means the model fits the data pretty well and captures nearly all the variance in the data (Table 5).
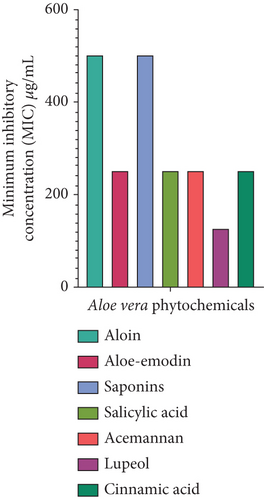
| ANOVA summary | Results |
|---|---|
| p value | < 0.0001 |
| p value summary | ∗∗∗ |
| R-squared | 0.9979 |
3.7. In Silico Analysis
The in silico analysis demonstrated that mutations in multiple genes contributed to antibiotic resistance. It focused on the structure of AmpC, which is known for its role in cephalosporin resistance through the overproduction of chromosomal cephalosporinase. Bioinformatics tools were used to investigate potential resistance-causing variants. The investigation revealed three key hotspot mutations in AmpC: T96I, G183D, and E247K. The T96I mutation involves the substitution of threonine with isoleucine at position 96, while G183D involves a glycine-to-aspartic acid change at position 183. The E247K mutation replaces glutamic acid with lysine at position 247. The three-dimensional structure of the AmpC protein (PDB ID 7FF0) was visualized using UniProt and AlphaFold to enhance the understanding of how these mutations affected antibiotic resistance (Figure S1). Docking was performed against the wild type and mutated AmpC structures using the cocrystal ligand alongside seven additional Aloe vera–derived phytochemicals: aloin, aloe-emodin, acemannan, saponin, lupeol, salicylic acid, and cinnamic acid selected based on virtual screening (Table 6).
| Code | Phytochemical | 2D structure | 3D structure | SMILES |
|---|---|---|---|---|
| PA-1 | Aloin | 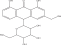 |
 |
C1=CC2=C(C(=C1)O)C(=O)C3=C(C2C4C(C(C(C(O4)CO)O)O)O)C=C(C=C3O)CO |
| PA-2 | Aloe-emodin | 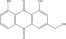 |
 |
C1=CC2=C(C(=C1)O)C(=O)C3=C(C2=O)C=C(C=C3O)CO |
| PA-3 | Acemannan |  |
 |
CC(=O)NC1C(C(C(OC1OC2C(OC(C(C2OC(=O)C)O)OC3C(OC(C(C3OC(=O)C)O)OC4C(OC(C(C4OC(=O)C)O)OC)CO)CO)CO)CO)OC5C(C(C(C(O5)CO)OC6C(C(C(C(O6)C(=O)[O-])OC7C(C(C(C(O7)CO)OC8C(C(C(C(O8)CO)OC)OC(=O)C)O)OC(=O)C)O)OC(=O)C)O)OC(=O)C)O)O |
| PA-4 | Saponin |  |
 |
CC1(C2CCC3(C(C2(CCC1OC4C(C(C(CO4)OC5C(C(C(CO5)O)O)O)OC6C(C(C(C(O6)CO)O)O)O)OC7C(C(C(C(O7)CO)O)O)OC8C(C(C(C(O8)CO)O)O)O)C)CCC91C3(CC(C2(C9CC(CC2)(C)C=O)CO1)O)C)C)C |
| PA-5 | Lupeol |  |
 |
CC(=C)C1CCC2(C1C3CCC4C5(CCC(C(C5CCC4(C3(CC2)C)C)(C)C)O)C)C |
| PA-6 | Salicylic acid | 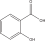 |
 |
C1=CC=C(C(=C1)C(=O)O)O |
| PA-7 | Cinnamic acid |  |
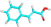 |
C1=CC=C(C=C1)C=CC(=O)O |
3.8. Pharmacokinetics, Drug-Likeness, and Toxicity
The key bioactive compounds of A. vera were examined, with LRF applied to assess physicochemical properties and drug-likeness. The compounds, PA-2, PA-6, and PA-7, showed no violations of Lipinski’s criteria, indicating that they possessed favorable drug-like qualities and the potential for oral bioavailability. PA-2 (aloe-emodin) emerged as a particularly promising phytochemical due to its low molecular weight (270.24 g/mol), favorable hydrophilicity (logP of 1.96), and optimal hydrogen bonding capacity (five acceptors and three donors). PA-6 (salicylic acid) was categorized as soluble according to both the estimated solubility (ESOL) and Silicos-it solubility models, with an ESOL logS of −2.5 and a Silicos-it logP of 0.74. Aloe-emodin was found to be moderately soluble, with an ESOL logS of −3.04 and a Silicos-it logP of 2.42. The compounds exhibited diverse logKp (cm/s) values in terms of permeability, which suggested variability in their ability to penetrate membranes. PA-2 and PA-6 showed moderately high permeability, with logKp values of −6.66 and −5.54, respectively, whereas lupeol demonstrated the highest permeability with a logKp value of −1.9, which suggested its stronger potential for absorption through biological membranes compared to the other compounds.
In terms of bioavailability, salicylic acid and cinnamic acid exhibited the highest scores of 0.85, indicating excellent potential for absorption and therapeutic effectiveness. The bioavailability score of 0.55 for aloe-emodin also suggested its favorable systemic distribution and therapeutic potential. The compounds varied in synthetic accessibility, with salicylic acid and cinnamic acid being the easiest to synthesize, having scores of 1 and 1.67, respectively. This analysis identified PA-2, PA-6, and PA-7 as the most promising candidates for further research based on their solubility, permeability, drug-likeness, bioavailability, and ease of synthesis (Table 7). Drug metabolism analysis, focusing on the cytochrome P450 enzyme system, revealed that PA-2 and PA-7 inhibited CYP1A2, while the other compounds did not inhibit the enzyme. Notably, PA-1, PA-3, PA-4, PA-5, and the cocrystal ligand were identified as substrates for CYP3A4, indicating a higher probability of being metabolized by this enzyme. None of the compounds inhibited CYP2C19, CYP2C9, CYP2D6, or CYP3A4, suggesting a lower risk of significant drug–drug interactions through these pathways.
| ADMET properties | PA-1 | PA-2 | PA-3 | PA-4 | PA-5 | PA-6 | PA-7 | CCL |
|---|---|---|---|---|---|---|---|---|
| MW | 418.39 | 270.24 | 1691.48 | 1223.35 | 426.72 | 138.12 | 148.16 | 328.2 |
| #H-bond acceptors | 9 | 5 | NA | 27 | 1 | 3 | 2 | 9 |
| #H-bond donors | 7 | 3 | NA | 15 | 1 | 2 | 1 | 2 |
| MR | 101.96 | 69.92 | NA | 285.71 | 135.14 | 35.42 | 43.11 | 68.7 |
| TPSA | 167.91 | 94.83 | NA | 422.05 | 20.23 | 57.53 | 37.3 | 167.48 |
| iLOGP | 1.86 | 1.96 | NA | 4.37 | 4.68 | 1.13 | 1.55 | 0.5 |
| XLOGP3 | −0.12 | 1.82 | NA | −2.67 | 9.87 | 2.26 | 2.13 | −2.96 |
| WLOGP | −1.04 | 1.21 | NA | −4.07 | 8.02 | 1.09 | 1.68 | −0.7 |
| MLOGP | −1.59 | 0.1 | NA | −6.13 | 6.92 | 0.99 | 1.9 | −2.31 |
| Silicos-IT logP | 0.18 | 2.42 | NA | −4.09 | 6.82 | 0.74 | 1.7 | −2.54 |
| Consensus logP | −0.14 | 1.5 | NA | −2.52 | 7.26 | 1.24 | 1.79 | −1.6 |
| ESOL logS | −2.46 | −3.04 | NA | −4.82 | −8.64 | −2.5 | −2.37 | −0.25 |
| Ali logS | −2.95 | −3.43 | NA | −5.64 | −10.22 | −3.1 | −2.54 | 0 |
| logKp (cm/s) | −8.94 | −6.66 | NA | −15.66 | −1.9 | −5.54 | −5.69 | −10.4 |
| CYP2D6 substrate | No | No | No | No | No | No | No | No |
| CYP3A4 substrate | Yes | No | Yes | Yes | Yes | No | No | Yes |
| CYP1A2 inhibitor | No | Yes | No | No | No | No | Yes | No |
| CYP2C19, 2C9, 2D6, 3A4 inhibitor | No | No | No | No | No | No | No | No |
| Bioavailability score | 0.55 | 0.55 | NA | 0.17 | 0.55 | 0.85 | 0.85 | 0.11 |
| PAINS #alerts | 0 | 1 | NA | 0 | 0 | 0 | 0 | 0 |
| Brenk #alerts | 0 | 0 | NA | 2 | 1 | 0 | 1 | 1 |
| Lead-likeness #violations | 1 | 0 | NA | 2 | 2 | 1 | 1 | 0 |
| Synthetic accessibility | 4.97 | 2.6 | NA | 10 | 5.49 | 1 | 1.67 | 4.38 |
- Note: PA-3 did not proceed in SwissADME due to a large number of atoms. CCL, cocrystal ligand.
The BOILED-Egg model provides valuable information about how compounds are absorbed and distributed. The model’s white region indicates favorable passive absorption in the gut, which suggests a high likelihood of oral bioavailability. The yellow area, which resembles an egg yolk, points to a strong possibility of crossing the blood–brain barrier (BBB), which implies potential involvement in central nervous system functions. The blue color identifies compounds that are substrates for P-glycoprotein (PGP+), a transporter that facilitates drug efflux, while red denotes compounds that are not substrates (PGP−). Among the key compounds, aloe-emodin fell within the white zone, indicating strong gastrointestinal absorption. Lupeol and salicylic acid were found within the yolk region, indicating a strong likelihood of crossing the BBB. In contrast, aloin, saponin, and cinnamic acid were positioned outside the egg, suggesting reduced capacity for both gastrointestinal absorption and BBB penetration (Figure S2).
The chosen phytochemicals were evaluated for acute toxicity and irritation characteristics based on multiple criteria, including inhalation, oral, and dermal toxicity, along with skin and eye irritation, using the StopTox tool (Table S2). All phytochemicals demonstrated no toxicity in terms of acute inhalation. However, salicylic acid exhibited both acute oral and dermal toxicity. Aloe-emodin and cinnamic acid were identified as toxic with regard to eye irritation and corrosion but were otherwise nontoxic across the other parameters. Cytotoxicity and hepatotoxicity evaluations were performed using the Protox 3.0 online platform, which estimated the potential toxicity of the chosen phytochemicals. According to these assessments, none of the compounds was anticipated to pose any major toxicity issues. Cinnamic acid, in particular, showed the most favorable outcomes, exhibiting minimal hepatotoxicity, neurotoxicity, and mutagenicity (Table S3). The ADMET and toxicity radar charts of the A. vera–derived phytochemicals are shown in Table S4.
3.9. Docking Studies
A molecular docking study was conducted to evaluate the interaction between the selected bioactive compounds from A. vera and the binding site of the AmpC protein in P. aeruginosa. Maestro was employed to dock the phytochemicals alongside the cocrystal ligand against both wild-type and mutant-type AmpC structures. To verify the docking outcomes, the structure was redocked with the cocrystal ligand, yielding an RMSD of 0.2002 Å for the redocked reference compound, which suggested a close structural match between the two positions in the AmpC binding site (Figure S3). All compounds exhibited different levels of interaction with the target protein in both the wild-type and mutated forms. In terms of binding affinity, compounds with more negative docking scores typically suggest a stronger attachment to the target site. Aloe-emodin, salicylic acid, and cinnamic acid showed significant docking scores of −7.97, −7.264, and −6.498 kcal/mol, respectively (Table 8), which highlighted their favorable binding interactions. The AmpC protein contained specific amino acids that demonstrated notable binding interactions with these compounds, such as F72, L66, A89, T132, R128, T133, L129, E74, and S88. The molecular visualization tools in PyMOL were employed to present the docking results for the cocrystal ligand and aloe-emodin, as illustrated in Figure 4, which highlights the interactions between the phytochemicals and the AmpC protein.
| Code | Phytochemical | Docking score | XP G score | Glide G score |
|---|---|---|---|---|
| 7FF0-ccl | Cocrystal ligand | −14.125 | −14.125 | −14.125 |
| PA-2 | Aloe-emodin | −7.97 | −7.97 | −7.97 |
| PA-6 | Salicylic acid | −7.264 | −7.264 | −7.264 |
| PA-7 | Cinnamic acid | −6.498 | −6.499 | −6.499 |
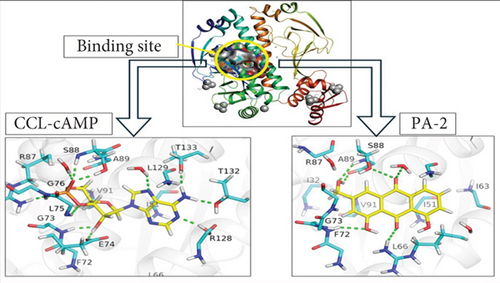
3.10. DFT Analysis of the Selected Ligands and Cocrystal Ligand
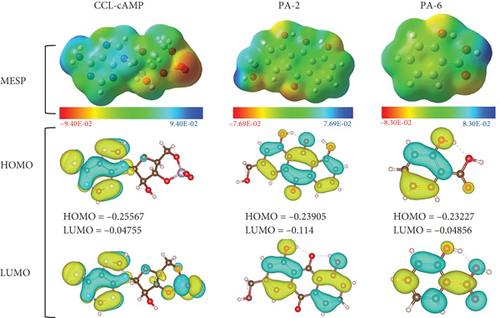
DFT analysis was performed to determine the electronic properties of the selected phytochemicals in the solvent phase (water) using the RB3LYP calculation technique. Salicylic acid and aloe-emodin were selected for DFT analysis based on the following criteria: ADMET profiling, bioactivity evaluation, and the interactions and binding affinities of the compounds. Of the ligands evaluated, aloe-emodin was the most reactive, with the smallest energy gap (0.12765 a.u.) and the highest softness value (7.835 eV), indicating that it was less stable but more flexible in terms of reactivity. The energy gap (ΔEGap) between HOMO and LUMO indicated both chemical stability and reactivity. While salicylic acid was more stable than aloe-emodin due to its larger energy gap (0.18371 a.u.), it had the highest electron affinity, as evidenced by the lowest LUMO energy (−0.04856 a.u.), making it more likely to absorb electrons. Aloe-emodin also had intermediate values for both HOMO and LUMO (Figure 5), suggesting a balanced reactivity profile compared to the higher electron-absorbing potential of salicylic acid (Table S5).
4. Discussion
P. aeruginosa is a highly virulent pathogen that poses significant risks, particularly as a cause of severe pulmonary infections. As part of a diverse and complex genus known for its role in various infections, P. aeruginosa represents a growing challenge due to its MDR nature. Our study underscores the need to explore novel phytochemicals from natural sources, such as A. vera, for their potential antimicrobial properties to combat these resilient infections. Antibiotics, including the antipseudomonal agents penicillins, carbapenems, cephalosporins, and fluoroquinolones, are generally effective against P. aeruginosa [40]. However, P. aeruginosa exhibits multidrug resistance and high virulence, thus posing a significant challenge to the effective treatment of infections. The susceptibility tests conducted in this study revealed high levels of AMR among the isolated organisms, especially to piperacillin/tazobactam and other beta-lactam agents with antipseudomonal activity; notably, 86.9% of the isolates demonstrated resistance to these drugs. In addition, first-line antibiotics, such as imipenem, cefepime, piperacillin/tazobactam, cefotaxime, and gentamicin, commonly employed to manage P. aeruginosa infections, are exhibiting diminished effectiveness [41, 42]. The growing resistance to these drugs contributes to increasing clinical complications and public health concerns. Research indicates that the frequent use of antibiotics drives genetic mutations in the pathogen, enabling it to acquire resistance and adapt more efficiently [19].
The analysis of the mutational resistome of P. aeruginosa can provide insights into its antibiotic-resistant genotype, thereby facilitating the development of targeted therapeutic strategies and improving the monitoring of antibiotic efficacy. Numerous genes and mutations are associated with elevated resistance levels and contribute to the formation of P. aeruginosa’s mutational resistome [43]. AmpC is recognized as a key contributor to antibiotic resistance mechanisms [44, 45]. In this study, we targeted three hotspot mutations in the AmpC gene, T96I, G183D, and E247K, which contribute to antibiotic resistance in P. aeruginosa [43, 46–48].
This study underscores the potential of A. vera–derived phytochemicals as antibacterial agents against drug-resistant infections and highlights the importance of further investigations into their clinical applications [23–25]. The pursuit of new bioactive compounds, particularly from natural sources, may offer promising solutions to combat resistance. Our study provides a detailed analysis of antibiotic resistance in P. aeruginosa isolates and explores the potential role of A. vera phytochemicals in addressing these challenging pathogens.
Multiple studies have consistently shown that A. vera holds considerable promise as a reservoir of novel bioactive molecules with potential medicinal applications. Its active constituents have shown potent antibacterial effects against a diverse array of pathogens [23–25]. A. vera comprises more than 75 unique compounds, which include vitamins, enzymes, minerals, sugars, anthraquinones (such as aloin and emodin), fatty acids (such as lupeol and campesterol), and hormones (auxins and gibberellins), along with other substances including salicylic acid, lignin, and saponins [25, 49]. These compounds exhibited promising antimicrobial properties through multiple mechanisms of action [50]. These phytochemicals may circumvent bacterial resistance mechanisms by targeting bacterial structures and pathways differently from traditional antibiotic therapies, thereby reducing the likelihood of cross-resistance. One of the key players is involved in the disruption of bacterial membranes. The disruption in bacterial membranes bypasses the conventional enzymatic resistance mechanism such as β-lactamase production or efflux pump activation. Several studies have demonstrated that compounds such as lupeol can compromise membrane integrity, leading to increased permeability and leakage of intracellular contents, which is often lethal to the bacteria [51]. Additionally, a few reported A. vera phytochemicals, such as aloe-emodin, have shown the potential to suppress the expression of efflux-related genes [52], thereby restoring the bacterial susceptibility to other antibiotics, particularly for P. aeruginosa [53]. One of the promising approaches is the inhibition of quorum sensing (QS), which is the main virulent factor for biofilm formation in P. aeruginosa, contributing to antibiotic resistance [54]. Certain Aloe vera components, such as acemannan and salicylic acid, have demonstrated anti-QS activity by impairing the bacteria’s ability to form protective biofilms [55]. Moreover, interference with DNA replication and protein synthesis has been reported. Aloe-emodin and cinnamic acid have been shown to intercalate into DNA or inhibit DNA gyrase activity, disrupting nucleic acid functions and leading to cell death without being affected by existing resistance enzymes [56]. Conclusively, these multifaceted actions—membrane disruption, efflux pump inhibition, QS suppression, and interference with DNA function—make Aloe vera phytochemicals compelling candidates for overcoming resistance mechanisms in MDR P. aeruginosa.
Based on the therapeutic significance of A. vera phytochemicals, the present study was centered on seven A. vera–derived phytochemicals—aloin, aloe-emodin, acemannan, saponin, lupeol, salicylic acid, and cinnamic acid. The antibacterial efficacy of these compounds against P. aeruginosa was assessed using the agar well diffusion assay. Cinnamic acid showed the strongest antibacterial activity with a 24-mm zone of inhibition, while acemannan exhibited a zone of inhibition of 22 mm. Acemannan, a polysaccharide, is known to boost immune responses, which could explain its antibacterial effects [57, 58]. Both aloe-emodin and lupeol exhibited zones of inhibition of 20 mm and 21 mm, respectively, suggesting that their antibacterial activity was moderate. Aloe-emodin, an anthraquinone, has demonstrated potential as an antimicrobial, antidiabetic, cytotoxic, cardioprotective, and bone-protective agent in in vitro research, as well as exhibiting anti-inflammatory and skin-protective effects in in vivo investigations [59–65]. The zone of inhibition for saponins was 18 mm, while aloin and salicylic acid had the lowest antibacterial activity, each showing a 15-mm zone of inhibition. These findings demonstrate that various A. vera phytochemicals are effective against MDR P. aeruginosa [23, 25]. Lupeol had the highest antibacterial potential, with an MIC of 125 μg/mL. Other compounds, including aloe-emodin, acemannan, and salicylic acid, may hinder bacterial growth by disrupting metabolic or enzymatic functions. The differences in MIC values likely stem from structural variations in the phytochemicals and their distinct modes of action within bacterial cells.
Future studies should investigate the unique interactions of these compounds with each other or in combination with established antibiotics to enhance their efficacy against resistant bacteria. Moreover, these phytochemicals offer a promising alternative source of novel antimicrobial agents, especially in the face of increasing antibiotic resistance. Several recent studies have employed extensive computational analysis to elucidate the inhibitory potential of the compounds for their use as promising therapeutic agents [66–68]. ADMET analysis and molecular docking studies were also conducted to further support these compounds’ antibacterial potential [33, 36]. The physicochemical and pharmacokinetic evaluations of A. vera’s key bioactive constituents, PA-2 (aloe-emodin), PA-6 (salicylic acid), and PA-7 (cinnamic acid), demonstrated favorable drug-like characteristics, suggesting strong oral bioavailability and pharmacological efficacy. Aloe-emodin showed advantageous properties, such as low molecular weight, well-balanced hydrogen bonding, moderate solubility, and reasonable permeability (logKp of −6.66), which positioned it as a promising therapeutic candidate. Cinnamic acid emerged as the most favorable of the compounds due to its minimal hepatotoxicity, neurotoxicity, and mutagenicity, making it an excellent subject for further investigation. The docking scores of aloe-emodin, salicylic acid, and cinnamic acid were −7.97, −7.264, and −6.498 kcal/mol, respectively, which suggest strong binding affinities. These compounds significantly interacted with amino acids, such as F72, L66, A89, T132, R128, T133, L129, E74, and S88, in the AmpC protein. These results provide important insights into the potential pharmaceutical and biomedical uses of these compounds, thereby establishing a foundation for selecting promising candidates for additional investigation and drug development. This study had some limitations, primarily due to its small sample size and the absence of in vivo experimentation to assess the efficacy and safety of A. vera extract.
To further validate the therapeutic potential of Aloe vera–derived phytochemicals, future investigations should focus on in vivo studies to assess pharmacodynamics, systemic efficacy, and toxicity profiles within a complex biological environment. These experiments are essential for determining the safety and therapeutic index of the compounds. Additionally, minimum bactericidal concentration (MBC) and time-kill assays should be employed to distinguish bactericidal effects from bacteriostatic activity, thereby providing a more comprehensive understanding of the antimicrobial potential. Comparative antibiotic testing is also recommended to benchmark the efficacy of these phytochemicals against conventional antibiotics and to evaluate their clinical relevance. Furthermore, elucidating the molecular mechanisms of action through advanced approaches such as transcriptomic profiling (e.g., RT-qPCR, RNA-Seq) will help to determine how these compounds influence gene expression and bacterial resistance pathways. Lastly, synergistic assays, involving the combination of these natural compounds with existing antibiotics, could uncover potential interactions that enhance antibacterial efficacy and may help to overcome existing resistance mechanisms. These multidimensional strategies will be instrumental in advancing the development of plant-derived antimicrobials into viable therapeutic agents.
5. Conclusion
The findings of the present study highlighted the potential of Aloe vera–derived phytochemicals as potent antibacterial agents to combat MDR P. aeruginosa. The in vitro analysis underscored cinnamic acid as the most potent antibacterial activity based on the zone of inhibition, whereas lupeol showed the lowest MIC value, indicating high potency at minimal concentrations. Simultaneously, the in silico analysis demonstrated the highest binding affinity for aloe-emodin in docking studies, suggesting a strong interaction with the AmpC β-lactamase target. Additionally, the ADMET analysis further supported the favorable pharmacokinetics and promising drug-likeness of aloe-emodin, positioning it as a suitable drug-like candidate for further pharmaceutical development against MDR P. aeruginosa. However, there is a need for further research into its clinical applications to combat drug-resistant bacterial infections.
Nomenclature
-
- ADME
-
- Absorption, distribution, metabolism, and excretion
-
- AMR
-
- Antimicrobial resistance
-
- BBB
-
- Blood–brain barrier
-
- CCL
-
- Cocrystal ligand
-
- CLSI
-
- Clinical and Laboratory Standards Institute
-
- DFT
-
- Density functional theory
-
- ESOL
-
- Estimated solubility
-
- HOMO
-
- Highest occupied molecular orbital
-
- LUMO
-
- Lowest unoccupied molecular orbital
-
- MDR
-
- Multidrug resistance
-
- MHB
-
- Mueller–Hinton broth
-
- MIC
-
- Minimum inhibitory concentration
-
- PGP
-
- P-glycoprotein
-
- WHO
-
- World Health Organization
-
- XDR
-
- Extensively drug-resistant
Ethics Statement
The study was approved by the ethical review committee of the University of Lahore, Pakistan, under Ref-IMBB/BBBC/24/1265 and conducted in accordance with the Declaration of Helsinki. This article does not contain any studies with human participants or animals performed by any of the authors. The study was conducted only on bacterial isolates.
Consent
The authors have nothing to report.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Conceived and designed the study: A.A., S.Y., and H.E.; performed the analysis: A.A., S.Y., M.U.K., Z.K., M.Al., M.Ab., A.E.A., K.A., and H.E.; funding acquisition: A.A.; investigation: A.A., S.Y., M.U.K., Z.K., M.Al., M.Ab., A.E.A., K.A., and H.E.; methodology: S.Y., M.U.K., Z.K., M.Al., M.Ab., A.E.A., K.A., and H.E.; supervision: A.A., S.Y., and H.E.; initial draft of the manuscript: M.U.K., Z.K., M.Al., M.Ab., A.E.A., and K.A.; critical review and editing of the manuscript: A.A., S.Y., and H.E. All authors read and approved the final manuscript. A.A. and S.Y. contributed equally to this work.
Funding
This work was funded by the Deanship of Graduate Studies and Scientific Research at Jouf University under grant No. DGSSR-2023-01-02388.
Acknowledgments
We would like to acknowledge the Deanship of Graduate Studies and Scientific Research at Jouf University, which supported this work.
Open Research
Data Availability Statement
The data used to support the findings of this study are included in the article.




