miR-21-5p Targets PIK3R1 to Regulate the NF-κB Signaling Pathway, Inhibiting the Invasion and Progression of Prolactinoma
Abstract
Prolactinomas (PRLs) are benign tumors with malignant characteristics that can invade the surrounding tissue structures and are challenging to treat. It has been reported that miR-21-5p expression in pituitary adenomas is correlated with tumor invasion and size. However, the mechanism of action of miR-21-5p in PRL remains unclear. Dysregulation of the phosphoinositide-3-kinase (PI3K) regulatory Subunit 1 pathway occurs frequently in cancer and plays an important role in tumor progression as an important component of the PI3K pathway. However, the role of PIK3R1 in PRL and its regulatory mechanism are unknown. In this study, we first explored the effect of miR-21-5p in PRL and then confirmed that PIK3R1 is a direct target of miR-21-5p using bioinformatics and cellular experiments. Subsequent in vitro experiments demonstrated that overexpression of PIK3R1 significantly attenuated the biological effects of miR-21-5p in PRL cells, such as promoting proliferation and invasion. Finally, we explored the mechanism by which PIK3R1 affects PRL progression and found that the inhibition of IκBa degradation by PIK3R1 impacts PRL progression via the miR-21-5p/PIK3R1/MMP pathway.
1. Introduction
Prolactinoma (PRL) is a tumor with endocrine function and is the most common tumor among all pituitary tumors [1]. Although PRL is a benign tumor, it has attracted much attention because of its distinct features, such as the anatomical location of the tumor and the symptoms produced by the tumor, including hyperprolactinemia that leads to amenorrhea, menstrual disorders, cessation of ovulation, male infertility, and sexual dysfunction in women. Some PRLs have invasive properties resulting in invasion of the surrounding tissues, which can lead to low vision, head pain, and can even be fatal [2]. Therefore, elucidating the pathogenesis of PRL is very important for its diagnosis and treatment. However, to date, studies on the molecular mechanism of PRL are very limited, and its molecular pathogenesis is controversial. The prevailing theory is that pituitary tumors are monoclonal tumors and that their proliferation stems from mutations in individual pituitary cells. However, it is unclear which pituitary cell mutations are more important for the local regulation of growth factors in the hypothalamus. Therefore, further research on the molecular mechanism of PRL is urgently needed.
miR-21 is overexpressed in many tumors, and reports have showed that miRNA-21 can play an important role in tumor proliferation and invasion in Type B malignant lymphoma, breast cancer, pancreatic cancer, and glioma [3–6]. Recently, studies have indicated that the interaction between miR-21-5p and long noncoding RNAs (lncRNAs) plays a significant role in the invasion and progression of various types of cancer [7]. It has also been reported that miR-21 expression is closely related to biological behaviors such as invasion and proliferation of pituitary adenomas [8, 9]. However, there is a lack of research on the role of miR-21 (miR-21-5p) in PRLs. In this study, we investigated the effect of miR-21 (miR-21-5p) on PRL at the cellular and animal levels.
As a major regulator of human cancer, phosphatidylinositol 3-kinase (PI3K) is a major signaling component downstream of growth factor receptor tyrosine kinase (RTK) [10]. In PRL, the peripheral prolactin receptor (PRLR), a member of the cytokine receptor superfamily, has been reported to transduce signals through the phosphoinositide 3-kinase-Akt (PI3K-Akt) or MAPK pathways to mediate changes in transcription, differentiation, and proliferation [11]. Numerous drugs tested in PRL cell lines have shown to exert inhibitory effects on tumor proliferation, mainly through the PI3K/AKT pathway [12]. Moreover, somatic mutations of the components involved in the activation of the PI3K pathway include mainly PIK3CA encoding the PI3K catalytic subunit p110α, PIK3R1 encoding the PI3K regulatory subunit p85α, and other genes encoding PI3K-related regulators, and it has been reported that PIK3R1 encodes p85a, followed by the action of downstream MMPs to play a tumor suppressor role [13–16]. The main mechanism underlying this effect is that after activation by PI3K/AKT, the transcription factor NF-κB is separated from IκB by the activation of IKK (IκB kinase), which phosphorylates IκBa and translocates to the nucleus to induce the expression of MMPs [17–19]. Therefore, based on previous studies, it is clear that it is additional studies on the mechanism of action of PIK3R1/MMPs in RPL are needed.
In this study, we selected MMP12 and MMP14 as downstream target genes of PIK3R1 to investigate whether PIK3R1 in PRL affects the progression of PRL through miR-21-5p/PIK3R1/MMPs.
2. Materials and Methods
Rat pituitary tumor cell line (Procell) contain the following: GH3, CL-0590 and MMQ, CL-0340.
2.1. PRL Samples
2.1.1. Patient Selection Criteria
The patients are selected based on the following criteria: (1) patients diagnosed with PRL; (2) patients who did not receive any radiotherapy, chemotherapy, or other related treatments before operation; (3) all patients had no other tumors except PRL; (4) patients with other underlying diseases, such as diabetes, hypertension, or severe heart disease, were excluded; (5) other family history of genetic diseases was negative. The relevant parameters of the enrolled patients are shown in Table 1.
| Parameters | Tumor invaded into the cavernous | |
|---|---|---|
| Yes | No | |
| Sex | ||
| M | 0 | 5 |
| F | 3 | 4 |
| Age (year) | ||
| 10–20 | 3 | 0 |
| > 20 | 1 | 8 |
2.2. Tissue Samples
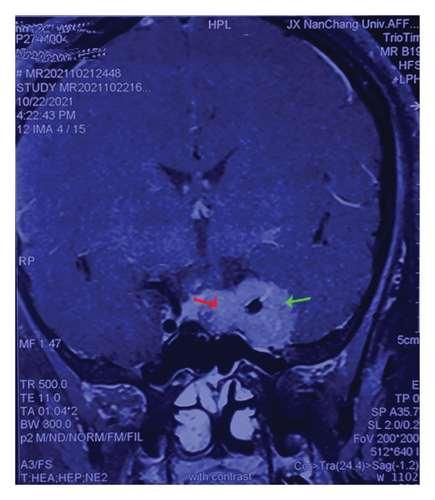
Twelve pairs of fresh PRL specimens were obtained after surgery from patients diagnosed with PRL (serum prolactin levels > 470 ng/mL) admitted to the First Affiliated Hospital of Nanchang University from January 2018 to October 2021. Each pair included two parts of tissue: intrasellar—juxtapituitary and cavernous sinus—away from the pituitary (away from the normal pituitary), which we considered to be more invasive. We isolated invasive PRLs (invasion part, Figure 1(a), green arrow) near pituitary tumor tissue (normal, Figure 1(a), red arrow), as tumors break through their capsule and invade the cavernous sinus farther away. The obtained tissues were stored immediately in a −80°C freezer. All enrolled patients were diagnosed with PRL according to the Chinese Consensus for the Diagnosis and Treatment of Pituitary PRL (2014 Edition) (specific diagnostic indicators had typical clinical manifestations, hyperprolactinemia, and imaging examination) [92]. Moreover, the case data of all patients were accurately and carefully recorded. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. Informed consent was obtained from the patients for the operation and tissue specimen collection, and relevant informed consent forms were signed before the operation.
2.3. Cell Culture and Subculture
For cell culture, MMQ and GH3 cells (Procell CL-0590) were cultured in Ham’s F-12K + 15% HS + 2.5% FBS + 1% medium (penicillin streptomycin solution) in a humidified incubator at 37°C and 5% CO2.
2.4. Cell Transfection
GH3 and MMQ cells in the logarithmic growth phase were inoculated in a 6-well plate at 1 × 105 cells/well and cultured overnight at 37°C and 5% CO2 saturated humidity. Two hours before transfection (CDNA, Youbao Biology, Hunan, China), the serum-free medium was used. The transfected cells were divided into the following groups: (1) normal cells: NC, (2) normal cells: NC mimics, and (3) miR-21-5p overexpression plasmid: miR-21-5p mimics. Finally, the cells were placed in an incubator at 37°C and a concentration of 5% CO2. After 6 h of culture, the medium was changed to normal medium and incubated for 24 h at 37°C with 5% CO2.
2.5. Transwell Assay
The collected cells, including those from the normal group and the transfection group, were resuspended, counted, diluted, and added to prepared 24-well plates. The transwell chamber was then placed on top of the plate. One hundred microliters of 1 mg/mL Matrigel matrix gel was added vertically to the center of the bottom of the upper chamber. After incubation, the target cells were inoculated, and the chamber was incubated again. Following incubation, the cells were fixed and stained. Finally, the invasive and migratory cells were observed and photographed under a microscope.
2.6. Real-Time Quantitative PCR
TRIzol reagent (Life Technologies) was used for RNA extraction. The synthesis of complementary DNA (cDNA) was performed with a Prime Script RT kit (Takara) and the first strand of Mir-X miRNA (Takara). The other procedures were the same as those in Part I. The miR-21-5p sequence was as follows: hsa miR-21-5p loop primer GTCGTATCCAGTGCAGG GT CCGAGGTATCGCA CTGGATCCAACCTC; F primer TGCGCTAGCTTA TCA GACTGA. The U6 and GAPDH primers were synthesized by Novozan Biotechnology Co., Ltd. qRT–PCR was used to determine the transcription level of some genes.
2.7. CCK-8
A CCK-8 kit (Sigma) was used to assess cell viability. The transfected GH3 and MMQ cells were seeded onto a 96-well plate at a density of 2 × 104 cells per well. Ten microliters (10 μL) of CCK-8 assay reagent was added to each well, and the plate was incubated for 2–4 h. The optical density (OD) values at 450 nm were subsequently measured using a microplate spectrophotometer for each well.
2.8. TUNEL Staining
The treated cells (GH3 slime, MMQ smear) were fixed in 4% paraformaldehyde (pH 7.4) solution at room temperature for 15 min. After being washed with PBS, centrifuged, and resuspended, the cells were incubated at room temperature for 15 min. The cells were incubated with bright red labeling mix on ice. The prepared TdT (Vazyme, A113-03) incubation buffer was added to the slides, and the slides were incubated for 60 min at 37°C (wrapped in aluminum foil paper in the dark). Finally, the core was restained and an image was acquired.
2.9. Tumorigenesis Assay
2.9.1. Animal Models
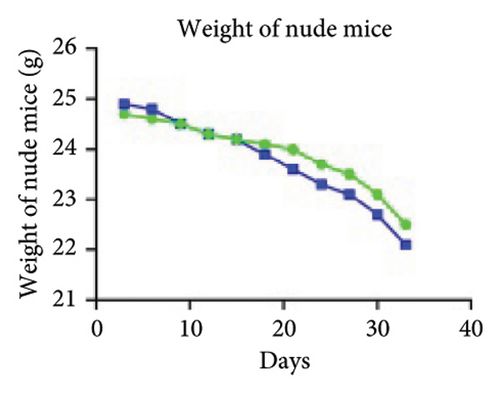
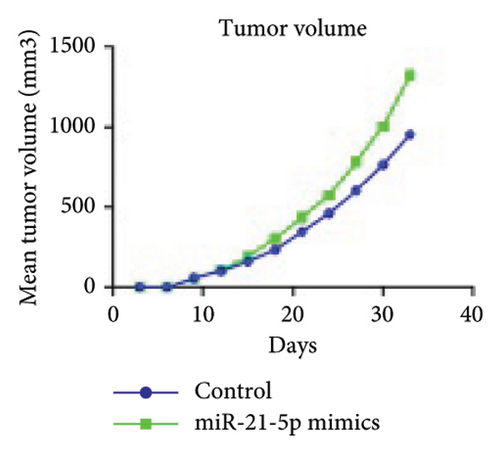
Firstly, nude mice were subjected to adaptive feeding for 7 days. For inoculation, 200 μL of cell suspension (containing approximately 1 × 107 cells) was injected after transfection and other treatments in the right axilla of nude mice, after wiping and disinfecting with alcohol cotton balls, and pinhole hemostasis was performed to continue cage feeding. The survival status and tumor size of the nude mice were observed and recorded, and after tumor emergence, the nude mice were weighed and recorded twice a week. The length and diameter of the tumors were measured with a Vernier caliper and recorded as a and b, and the tumor volume calculation formula V = ab2/2 was used.
When the tumor volume reached 60 mm3, the nude mice were randomly divided into two groups, with three nude mice in each group: (1) the normal group and (2) the miR-21-5p overexpression group.
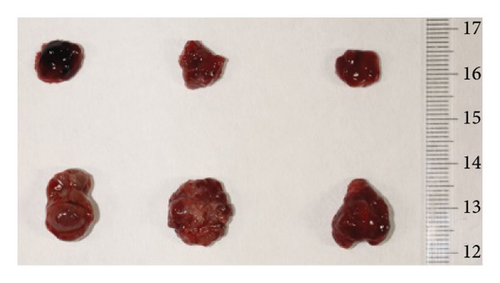
On the 33rd day of the experiment, all the nude mice were sacrificed via cervical dislocation. The tumors were removed, neatly placed in a monochromatic background according to groups, and photographed with a ruler. Then, half of the tumor tissues were fixed in 4% paraformaldehyde and half were cryopreserved for relevant index detection.
2.10. Immunohistochemistry
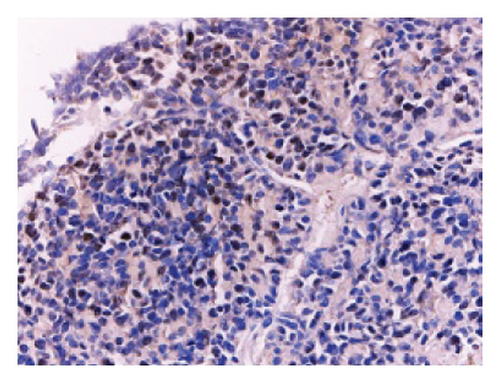
The tissue blocks were analyzed using standard immunohistochemical experimental procedures for gradient alcohol for tissue dehydration, tissue transparency, redehydration, wax immersion, embedding, sectioning and baking, dewaxing, antigen retrieval, blocking endogenous peroxidase, serum blocking, spiking primary antibody, spiking secondary antibody, spiking chromogen counterstaining, dehydration, mounting, and microscopic photography (not repeated), and finally, OD analysis of immunohistochemical photographs was performed using IPP6.0 software. The dilution concentration of primary antibody against Ki67 was 1:100.
2.11. Luciferase Reporting Assay
The groups were as follows: (1) normal 293T control cells, (2) WT-PIK3R1 3′UTR reporter plasmid + NC mimics, (3) WT-PIK3R1 3′UTR reporter plasmid + miR-21-5p mimics, (4) MUT-PIK3R13′UTR reporter plasmid + NC mimics, and (5) MUT-PIK3R1 3′UTR reporter plasmid + miR-21-5p mimics. After the cultured cells were exhausted, 200 μL of reporter cell lysate was added. After full lysis and centrifugation at 12,000 RPM for 3 min, the supernatant was collected for analysis. The firefly luciferase detection reagent and Renilla luciferase detection buffer were dissolved and incubated at room temperature. Renilla luciferase detection substrate (100×) was placed in an ice bath or an ice box for use. An appropriate amount of Renilla luciferase detection buffer was added to each sample (100 μL), and Renilla luciferase detection substrate (100×) was added at a ratio of 1:100 to prepare a Renilla luciferase detection working solution. Fifty microliters of sample was added, 100 μL of firefly luciferase detection reagent was added, the mixture was mixed well, and the RLU was determined. One hundred microliters of Renilla luciferase was added to the working solution, and the RLU was determined after mixing. With firefly luciferase as an internal reference, the RLU value determined by Renilla luciferase was divided by the RLU value determined by firefly luciferase.
2.12. Western Blot Analysis
The cells were collected on ice, lysed with lysis buffer, and centrifuged at 12, 000 × g for 30 min, after which the supernatant was retained. Protein lysates were loaded onto SDS-GEL, separated via SDS–PAGE and transferred to PVDF membranes. After blocking with 20% (PBS-Tween) and 5% BSA for 1 h, the film was incubated overnight with the following primary antibodies: anti-GAPDH (Hangzhou Xian Zhi Biology Co., Ltd., AB-P-R001), rabbit IKB-a (39 kDa Affinity AF5002), rabbit p-IKB-a (39 kDa Affinity AF3239), rabbit PI3K (85 kDa Affinity AF6241), rabbit MMP14 66 kDa AF0212);, and rabbit MMP12 (54 kDa DF7686), followed by washing in Tris-buffered saline with Tween 20 (10 min, 3 washes). Then, the sections were incubated with the appropriate secondary antibody (1:5000; Wuhan Baishi Biological Engineering Co., Ltd., BA1054). The signal was observed using enhanced chemiluminescence reagent (Tianjin Hanzhong Photographic Materials Factory), and density analysis was performed using ImageJ (National Institutes of Health).
2.13. Principal Component Analysis (PCA) of the Microarray Data
PCA was performed using R package stats (version 3.6.0) for GSE32191, which was standardized to determine the reproducibility of the data.
2.14. Heatmap, Volcano Plot, and Differential Expression Gene Analysis
Heatmaps and volcano plots were generated using the R package “heatmap” with “ggplot2.” Analysis of differentially expressed genes (DEGs) was performed on the dataset using the R package “limma.” Genes with p < 0.05 were considered differentially expressed.
2.15. Venn Diagrams
The online website Draw Venn Diagrams (https://bioinformatics.psb.ugent.be/webtools/venn/) was used for Venn diagrams.
2.16. Protein-Protein Interaction (PPI) Network Diagram Analysis
The STRING database was used to draw PPIs. Cytoscape software (version 3.9.1) and cytoHubba were used to analyze the hub genes.
2.17. miRNA Prediction Platform for Prediction of Target Genes for miR-21-5p
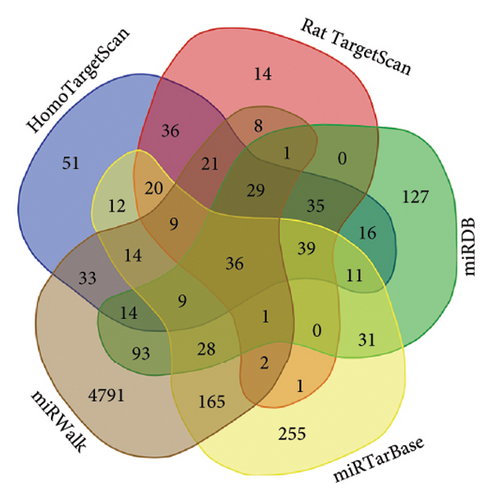
The online web target prediction platforms used included miRTarBase (miRTarBase [Mayanlab.cloud]), miRWalk (Home-miRWalk [Fig. uni-heidelberg.de]), miRDB (miRDB-MicroRNA Target Prediction Database), and TargetScan (TargetScanHuman 7.1). The TargetScan platform was used to predict data for two species, humans and mice, and a total of five sets of data were obtained. The GEO database GSE32191 microarray data were downloaded (the annotation platform of GSE32191 is GPL2895), which contained 8 invasive samples and 5 noninvasive samples. DEGs were analyzed between the invasive and noninvasive groups.
2.18. Statistical Analysis
Statistical analysis was performed using SPSS software (version 20.0; IBM Company). Correlation analysis was performed via the Spearman correlation. Student’s t test and one-way analysis of variance were used to calculate differences between 2 or more groups. p < 0.05 was considered statistically significant (p < 0.05, p < 0.01, and p < 0.001; ns = not significant).
3. Results
3.1. miR-21-5p Expression Is Significantly Higher in PRL Tissues Than in Normal Tissues, and Its Expression Is Positively Correlated With Invasiveness
In this study, we performed preoperative magnetic resonance imaging (MRI) on the included patients. All samples were selected and labeled as shown in Figures 1(a): red arrows indicate the juxtapituitary regions, areas of weak invasiveness, or no invasiveness; green arrows indicate the distal pituitary tumors and areas of strong invasiveness. The relevant parameters of the included patients are shown in Table 1.
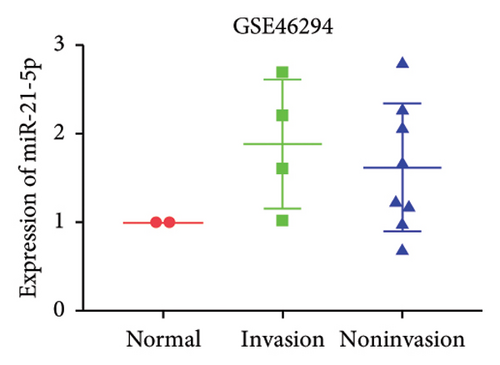
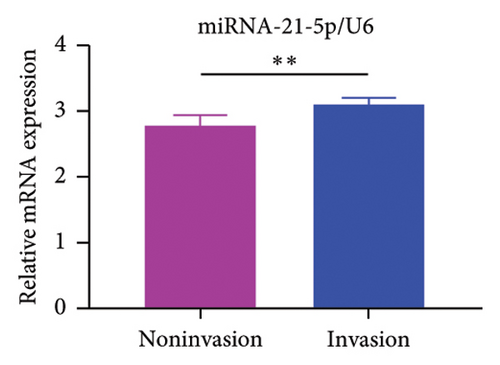
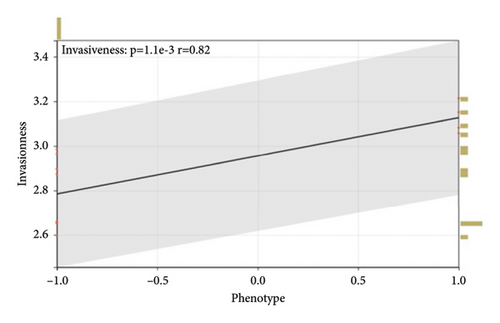
GEO2R analysis of miR-21-5p differential expression in invasive versus noninvasive pituitary tumors in the GSE46294 dataset revealed that miR-21-5p expression was significantly higher in invasive PRLs than in noninvasive PRLs (Figures 1(b)). A PCR assay of clinical samples revealed (Figures 1(c)) that miR-21-5p expression in invasive PRL tissues invading the cavernous sinus was significantly higher than that in noninvasive tissues, and the results were consistent with the GEO data. To elucidate the correlation between miR-21-5p and PRL invasiveness, we defined invasive PRL (Y value set as 1) in 4 of the 12 samples in which the tumor broke through its capsule and invaded the cavernous sinus and noninvasive PRL (Y value set as −1) in the other 8 samples in which the PRL did not invade the cavernous sinus, and the correlation between the expression of miR-21-5p, and the invasive phenotype was calculated using the Spearman correlation statistics. The results revealed that the expression of miR-21-5p was positively correlated with PRL invasiveness, with a correlation of R = 0.82 and p = 0.0011, as shown in Figure 1(d).
3.2. miR-21-5p Promotes the Progression of PRL In Vitro and In Vivo
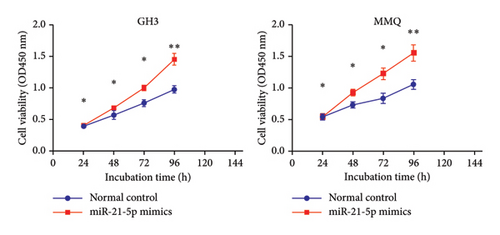
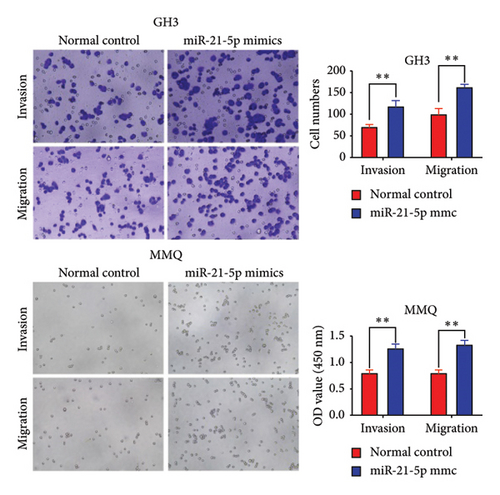
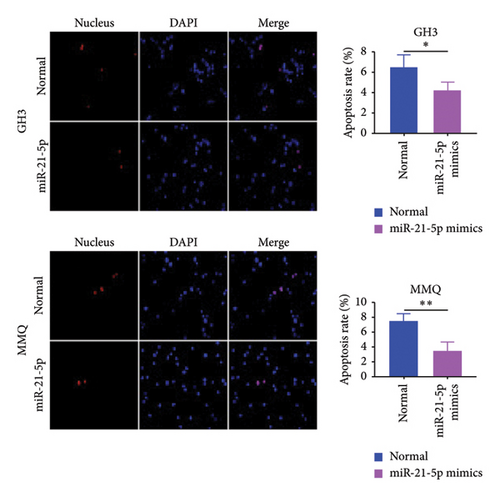
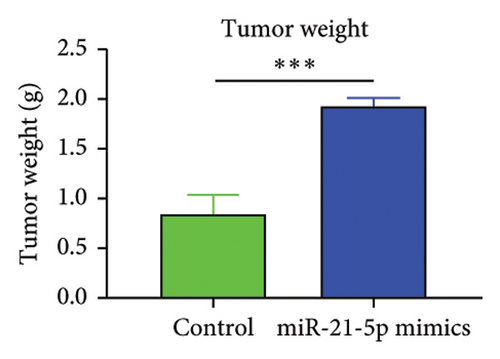
To investigate the effect of miR-21-5p on the biological behavior of PRL cells, we overexpressed miR-21-5p in PRL cells and detected the effect of overexpressing miR-21-5p on the biological behavior of PRL cells using CCK8, transwell, and TUNEL assays. The results revealed that the proliferation (as shown in Figure 2(a)), invasion, and migration (as shown in Figure 2(b)) of PRL cells were significantly increased, and that apoptosis was attenuated after miR-21-5p overexpression (as shown in Figure 2(c)). PRL cells can promote PRL cell proliferation, invasion, and migration and inhibit apoptosis. Moreover, miR-21-5p promoted the proliferation and invasion of PRL tumors in vivo (as shown in Figure 3).
3.3. PIK3R1 Is a Hub Gene for PRL Invasiveness
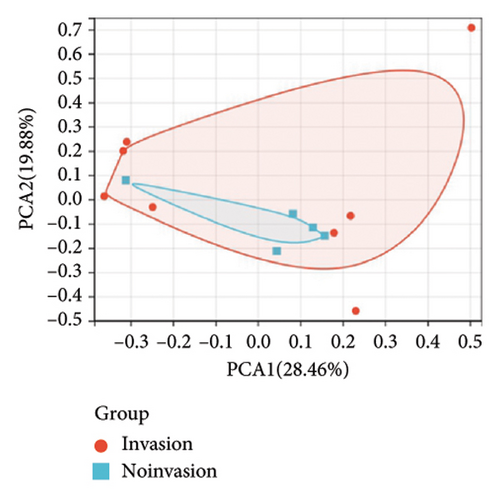
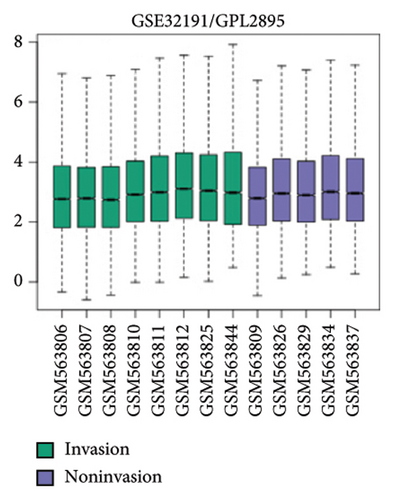
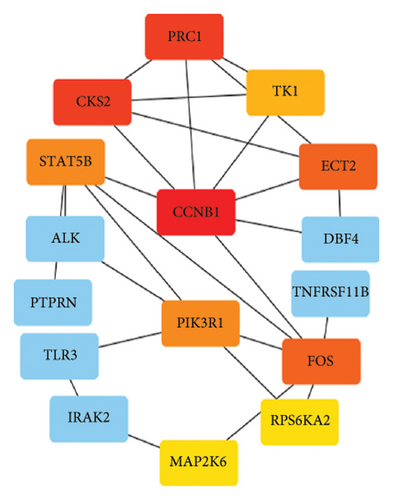
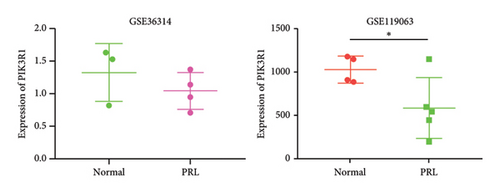
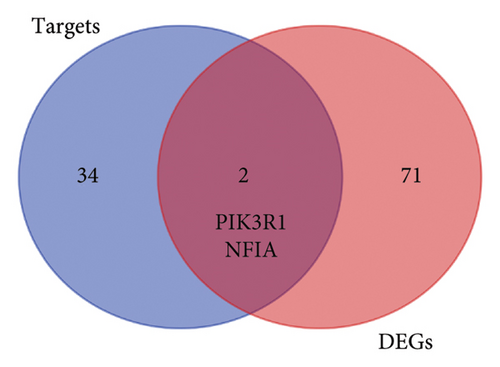
To further explore the downstream mechanism of miR-21-5p, we first analyzed GSE32191 to obtain the hub gene set of PRL invasiveness (Figures 4(a) and 4(b) PCA and box plots for GSE32191, respectively), and these results demonstrated the good reproducibility of the GSE32191 data. GSE32191 was used for differential gene expression (DEG) analysis, and the top 50 DEGs were used for PPI network analysis (Figure 4(c)), followed by network mapping of the top 50 DEGs using cytoHubba interactions (Figure 4(d)). These data indicate that PIK3R1 is the s-hub gene for PRL invasiveness. The GSE36314 and GSE119603 datasets subsequently verified that PIK3R1 expression was significantly higher in PRL patients than in normal controls, as shown in Figure 4(e). We used the online miRNA target prediction platforms miRTarBase miRWalk, miRDB, and TargetScan to predict the set of target genes for miR-21-5p, as shown in Figure 4(f). Subsequently, a Venn diagram of the predicted target genes against the differential genes of GSE32191 revealed that PIK3R1 and NFIA were the target genes, as shown in Figure 4(g). These results indicate that PIK3R1 is a key gene for PRL invasiveness.

3.4. PIK3R1 Is a Direct Target of miR-21-5p and Inhibits PRL Proliferation
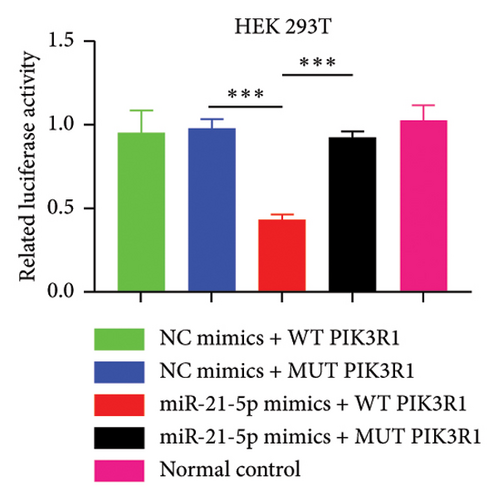
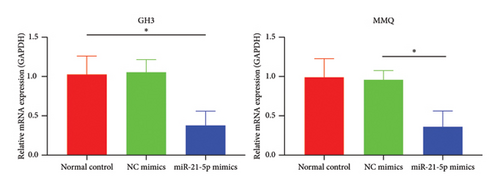
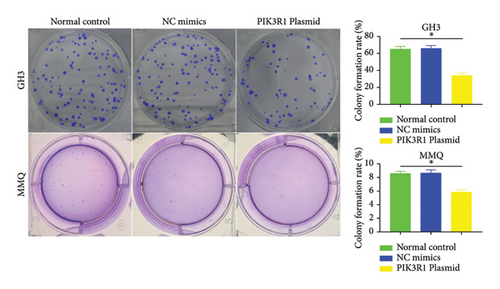
Recently, a report indicates that miR-21 targets PIK3R1 to influence the progression of various tumors. However, there are currently no reports on its role in PRL [20]. We used the prediction platform TargetScan to predict base complementary pairing of miR-21-5p with the 3′UTR of PIK3R1, as shown in Figure 5(a). Then, dual-luciferase assays were performed to further validate whether PIK3R1 is a target of miR-21-5p, and dual-luciferase reporter assays were performed after cotransfection of miR-5p 21-mimics and WT-PIK3R1 3′UTR/MUT-PIK3R1 3′ UTR in 293T cells. The results revealed that the luciferase activity of the PIK3R1-3′UTR wild-type reporter plasmid was significantly inhibited after transfection with the miR-21-5p mimics, but there was no significant change in the luciferase activity of the PIK3R1-3′UTR mutant reporter plasmid (Figure 5(b)). Thus, these results indicate that PIK3R1 is a direct target gene of miR-21-5p. Subsequent cell experiments revealed that the overexpression of miR-21-5p in PRL cells inhibited PIK3R1 expression at both the mRNA and protein levels, as shown in Figures 5(c) and 5(d). Cell cloning assays revealed that PIK3R1 significantly inhibited the proliferation and progression of PRL cells (Figure 5(e)).
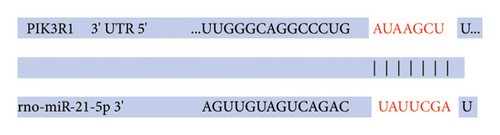
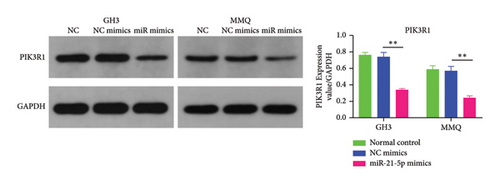
3.5. PIK3R1 Inhibits IκBa Degradation, Affecting the Progression of PRL via the miR-21-5p/PIK3R1/MMP Pathway
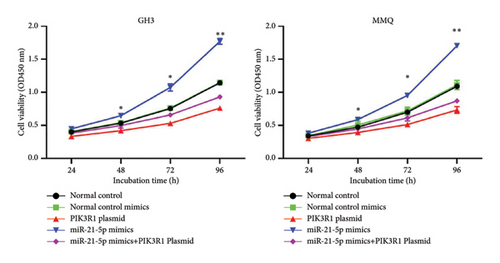
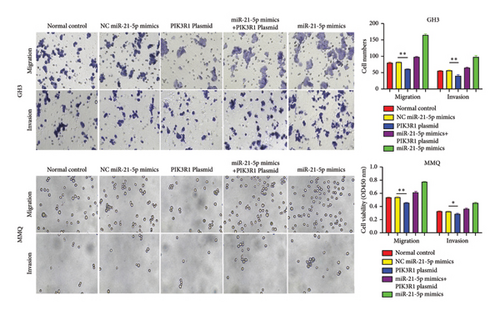
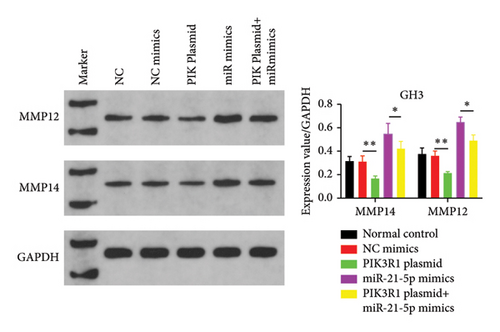
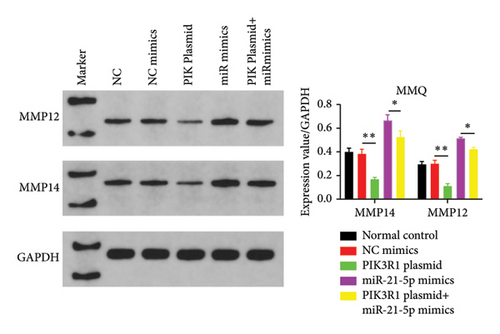
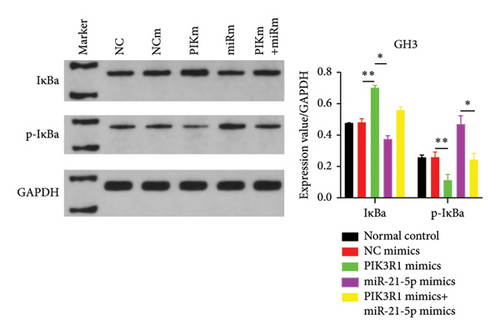
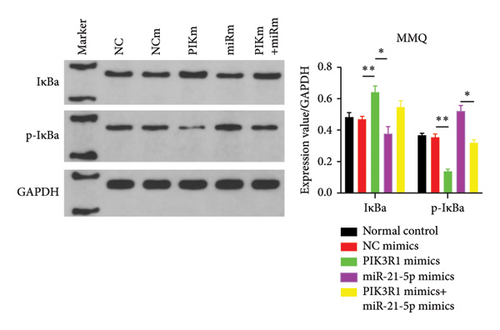
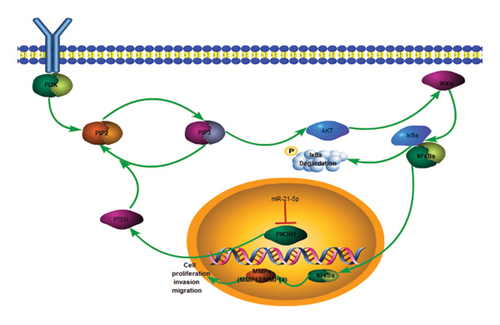
On the basis of the previous experimental results, we transfected miR-21-5p mimics and PIK3R1 plasmids into GH3 and MMQ cells and then performed CCK8 and transwell assays to determine whether overexpression of PIK3R1 can reduce the ability of miR-21-5p to promote the proliferation, invasion, and migration of PRL-producing cells. CCK8 and transwell assays revealed that the proliferation, invasion, and migration abilities of the cells in the miR-21-5p mimics + PIK3R1 plasmid group were significantly lower than those in the miR-21-5p mimics + empty transfer group, and there was no significant difference between the two groups and the NC mimics + empty transfer group. As shown in Figures 6(a) and 6(b), these results demonstrated that the overexpression of PIK3R1 can inhibit the promoting effect of miR-21-5p in PRL cell proliferation, invasion, and migration. As described above, we transfected miR-21-5p mimics and PIK3R1 plasmids into GH3 and MMQ cells and then examined whether overexpression of PIK3R1 could inhibit the effects of miR-21-5p intervention on genes related to PIK3R1 downstream genes in PRL cells. The results revealed that IκBa phosphorylation was significantly lower and that the total amount of IκBa was significantly greater in the PIK3R1 plasmid group than in the other groups (as shown in Figures 7(a) and 7(b)). Moreover, the expression of MMP12/MMP14 was significantly lower in the PIK3R1 plasmid group than in the other groups (as shown in Figures 7(c) and 7(d)). PIK3R1 inhibits IκBa degradation, thereby affecting the progression of PRL via the miR-21-5p/PIK3R1/MMPs pathway (as shown in Figure 7(e)).
4. Discussion
At present, the treatment of PRL remains controversial, and consensus guidelines recommend dopamine agonists (DAs) as first-line treatments [21, 22], and surgery is indicated in patients who are resistant to DAs or cannot tolerate the side effects, or in cases of cystic fibroadenoma and persistent tumor bleeding with visual impairment [23–27]. In 2006, the Pituitary Society revised the guidelines, stating that surgery can be used as a first-line treatment with respect to patients’ wishes (some patients cannot receive long-term DAs, even if they do not experience resistance or intolerance) [28]. However, there are still several limitations and risks in surgical treatment, such as whether the tumor can be completely removed by surgery and the possible risks and complications during and after surgery. Therefore, further studies on the mechanism of PRL occurrence and progression are necessary.
miR-21-5p has been reported to play an important role in the development and progression of a variety of tumors and diseases, including gastric cancer, lung cancer, rectal cancer, glioma, and melanoma [29–33]. Tang et al. reported that miR-21-5p plays an important role in the progression of lung cancer [34]. Zhang et al. reported that miR-21-5p plays an important role in the proliferation and progression of acute myeloid leukemia [35]. Li et al. reported that miR-21-5p plays an indispensable role in the progression of osteosarcoma [36], and it has also been shown that miR-21 expression in pituitary adenomas is closely related to tumor invasion and size [37]. However, whether miR-21-5p promotes or inhibits PRL biobehavior has not previously been reported.
In this study, both the GEO database data and the PCR results of the clinical samples revealed that the expression of miR-21-5p in the invasive part of PRL was significantly greater than that in the normal and noninvasive parts. In addition, the expression level of miR-21-5p was positively correlated with the invasiveness of PRL, and the more invasive the PRL was, the higher the expression level of miR-21-5p was.
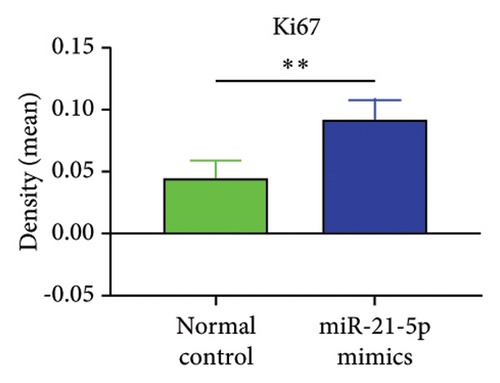
The in vitro data revealed that miR-21-5p promoted the proliferation, migration and invasion of PRL cells and reduced their degree of apoptosis. In vivo experiments also demonstrated that miR-21-5p significantly promoted tumor growth in vivo without significant differences in body weight in nude mice (indicating that nude mice are similar under physiological conditions). Related data on Ki67 also suggest that miR-21-5p is able to promote PRL invasiveness [38, 39]. To identify the target of miR-21-5p in PRL, we first searched for potential target gene sets through a target prediction platform, and DEG analysis was subsequently performed on the GSE32191 invasive and noninvasive samples to identify DEGs. PIK3R1 was ultimately identified as the target gene. Dual-luciferase assays revealed that PIK3R1 is a direct target of miR-21-5p, which can directly act on PIK3R1 and inhibit the expression of PIK3R1 mRNA and protein. Cloning experiments further demonstrated that PIK3R1 can significantly inhibit the proliferation and progression of PRL cells.
Dysregulation of the PI3K pathway is well known to play an important role in the progression of numerous tumors including PRL [40, 41]. The role of PIK3R1, one of the major components of somatic mutations in the PI3K pathway, in the PI3K pathway is critical [42, 43]. Therefore, it is necessary to study the role of PIK3R1 in PRL and its related mechanisms.
PIK3R1 encodes P85a, which stabilizes PTEN, and PTEN induces the dephosphorylation of PIP3 to PIP2, which plays a role in tumor suppression [15, 44]. PI3K recruits PDK1 and AKT proteins to the plasma membrane, causing PDK1 to phosphorylate threonine 308 (T308) at position 308 of the AKT protein, resulting in partial activation of AKT, whereas at rest, IκBa binds to two subunits of NFκB (p6 and, p50) in an inactivated state and is present in the cytoplasm. When upstream genes activate AKT, AKT activates IKK, and activated IKK undergoes ubiquitination and phosphorylation to degrade IκBα, ultimately activating the two subunits of NF-κB and further transferring them from the cytoplasm into the nucleus to promote the expression of MMPs, which in turn promotes enhanced biological behaviors such as cell proliferation, invasion, and migration [16, 45, 46].
In this study, we found that PIK3R1 overexpression promoted decreased IκBa phosphorylation and increased IκBa; then, IκBa bound to more NFκB subunits to inactivate NFκB, and the cancer-promoting effect of NFκB was significantly reduced, which is consistent with the findings of previous studies. Thus, our data demonstrate that PIK3R1 induces reduced expression of MMPs (MMP12/MMP14) by reducing IκBa phosphorylation and ultimately leads to diminished proliferation, invasion, and migration of PRL cells.
In summary, the mechanism of the inhibitory effect of PIK3R1 in PRL is as follows: PIK3R can stabilize PTEN, followed by PTEN, to increase PIP3-to-PIP2 conversion, and attenuate the effect of the PI3K-PIP2-PIP3-AKT pathway [15, 44, 47]; subsequently, IκBa phosphorylation is reduced, IκBa content is increased (IκBa will bind to more NFκB subunits to inactivate NFκB), and the cancer-promoting effect of NFκB is significantly reduced [16, 45, 46]), resulting in reduced expression of MMPs (MMP12/MMP14) to achieve a tumor suppressor effect. In conclusion, our data demonstrate that PIK3R1 reduces IκBa phosphorylation and impacts PRL progression via the miR-21-5p/PIK3R1/MMP pathway (Figure 7).
In summary, the interaction of miR21-5p with PIK3R1 triggers IκBa phosphorylation, which is reduced by PIK3R1 and affects the progression of PRL via the miR-21-5p/PIK3R1/MMP pathway. This pathway is therefore an important potential target to prevent and treat PRL.
Ethics Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee of the First Affiliated Hospital of Nanchang University. All animal experimental protocols were performed in accordance with relevant guidelines, and all methods were reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) and approved by the Use Committee for Animal Care at the First Affiliated Hospital of Nanchang University. All experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from all individuals or their guardians. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The studies involving human participants (informed consent was obtained from guardians for all minor subjects) were reviewed and approved by the First Affiliated Hospital of Nanchang University. The patients/participants provided written informed consent to participate in this study. The animal study was reviewed and approved by the First Affiliated Hospital of Nanchang University.
Consent
This work received consent to publish from all authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Min de Li performed the experiments and generated the data. Min de Li, Juan Yang, Shang Si Chen, and Xiao Wu analyzed the data. Min de Li and Shang Si Chen designed the experiments. Shang Si Chen and Min de Li wrote the manuscript. Min de Li and Juan Yang revised the manuscript. All the authors contributed to the article and approved the submitted version. Min de Li and Juan Yang contributed equally.
Funding
No funds were received.
Acknowledgments
The authors have nothing to report.
Open Research
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GEO Repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46294, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32191, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36314, and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119063). For additional data, please contact the corresponding author (email: [email protected]).